-
PDF
- Split View
-
Views
-
Cite
Cite
John J. Flynn, John A. Finarelli, Sarah Zehr, Johnny Hsu, Michael A. Nedbal, Molecular Phylogeny of the Carnivora (Mammalia): Assessing the Impact of Increased Sampling on Resolving Enigmatic Relationships, Systematic Biology, Volume 54, Issue 2, April 2005, Pages 317–337, https://doi.org/10.1080/10635150590923326
Close - Share Icon Share
Abstract
This study analyzed 76 species of Carnivora using a concatenated sequence of 6243 bp from six genes (nuclear TR-i-I, TBG, and IRBP; mitochondrial ND2, CYTB, and 12S rRNA), representing the most comprehensive sampling yet undertaken for reconstructing the phylogeny of this clade. Maximum parsimony and Bayesian methods were remarkably congruent in topologies observed and in nodal support measures. We recovered all of the higher level carnivoran clades that had been robustly supported in previous analyses (by analyses of morphological and molecular data), including the monophyly of Caniformia, Feliformia, Arctoidea, Pinnipedia, Musteloidea, Procyonidae + Mustelidae sensu stricto, and a clade of (Hyaenidae + (Herpestidae + Malagasy carnivorans)). All of the traditional “families,” with the exception of Viverridae and Mustelidae, were robustly supported as monophyletic groups. We further have determined the relative positions of the major lineages within the Caniformia, which previous studies could not resolve, including the first robust support for the phylogenetic position of marine carnivorans (Pinnipedia) within the Arctoidea (as the sister-group to musteloids [sensu lato], with ursids as their sister group). Within the pinnipeds, Odobenidae (walrus) was more closely allied with otariids (sea lions/fur seals) than with phocids (“true” seals). In addition, we recovered a monophyletic clade of skunks and stink badgers (Mephitidae) and resolved the topology of musteloid interrelationships as: Ailurus (Mephitidae (Procyonidae, Mustelidae [sensu stricto])). This pattern of interrelationships of living caniforms suggests a novel inference that large body size may have been the primitive condition for Arctoidea, with secondary size reduction evolving later in some musteloids. Within Mustelidae, Bayesian analyses are unambiguous in supporting otter monophyly (Lutrinae), and in both MP and Bayesian analyses Martes is paraphyletic with respect to Gulo and Eira, as has been observed in some previous molecular studies. Within Feliformia, we have confirmed that Nandinia is the outgroup to all other extant feliforms, and that the Malagasy Carnivora are a monophyletic clade closely allied with the mongooses (Herpestidae [sensu stricto]). Although the monophyly of each of the three major feliform clades (Viverridae sensu stricto, Felidae, and the clade of Hyaenidae + (Herpestidae + Malagasy carnivorans)) is robust in all of our analyses, the relative phylogenetic positions of these three lineages is not resolvable at present. Our analyses document the monophyly of the “social mongooses,” strengthening evidence for a single origin of eusociality within the Herpestidae. For a single caniform node, the position of pinnipeds relative to Ursidae and Musteloidea, parsimony analyses of data for the entire Carnivora did not replicate the robust support observed for both parsimony and Bayesian analyses of the caniform ingroup alone. More detailed analyses and these results demonstrate that outgroup choice can have a considerable effect on the strength of support for a particular topology. Therefore, the use of exemplar taxa as proxies for entire clades with diverse evolutionary histories should be approached with caution.The Bayesian analysis likelihood functions generally were better able to reconstruct phylogenetic relationships (increased resolution and more robust support for various nodes) than parsimony analyses when incompletely sampled taxa were included. Bayesian analyses were not immune, however, to the effects of missing data; lower resolution and support in those analyses likely arise from non-overlap of gene sequence data among less well-sampled taxa. These issues are a concern for similar studies, in which different gene sequences are concatenated in an effort to increase resolving power.
In this study, we expanded sampling of both taxa and characters beyond all previous analyses of primary molecular phylogenetic data for the Carnivora (Mammalia) (Appendix 1). Monophyly of the Carnivora and many major carnivoran subclades has been well established for some time, based on morphological, molecular, and integrative phylogenies (e.g., Flynn and Galiano, 1982; Flynn et al., 1988, 2000; Wyss and Flynn, 1993; Flynn and Nedbal, 1998; Bininda-Emonds et al., 1999; Yoder et al., 2003; see Flynn and Wesley-Hunt, 2005). Within the Carnivora a bifurcation between two monophyletic clades, the Caniformia and the Feliformia, has been robustly supported, as has the monophyly of “suprafamilial” groups such as the Arctoidea, Pinnipedia, and Musteloidea. In addition, all of the traditional “family”-level groupings (e.g., Wozencraft, 1993) appear to represent monophyletic clades, with the exception of the Viverridae and Mustelidae. Interrelationships among the living feliform “families” (the Feloidea: “Viverridae,” Herpestidae, Hyaenidae, Felidae), as well as of several enigmatic taxa (i.e., Nandinia [African palm civet; traditionally placed in the Viverridae] and Madagascar's feloids [traditionally placed in three or more families, Viverridae, Herpestidae, Felidae]), have long been ambiguous. Recent molecular phylogenies of the feliform carnivorans have yielded increased resolution and several strongly supported clades, such as a monophyletic clade of the Malagasy carnivorans that is the sister-group to Herpestidae sensu stricto (s.s.), the Hyaenidae as sister-group to this clade, and Nandinia as nearest outgroup to all other extant feloids (Yoder et al., 2003; Yoder and Flynn, 2003; Gaubert and Veron, 2003). However, ambiguity remains as to the exact interrelationships near the base of the feloid radiation (i.e., is Felidae or Viverridae [s.s.] more closely related to the Malagasy carnivoran-Herpestidae-Hyaenidae clade?). Within the Caniformia, most remaining ambiguity centers on the precise interrelationships among various family-level groups within the Arctoidea (Ledje and Árnason, 1996a, 1996b; Flynn and Nedbal, 1998; Flynn et al., 2000). In addition, recent molecular phylogenies are at odds with morphologic hypotheses over the relationships of skunks and stink badgers. Traditionally, these taxa have been placed within the Mustelidae (Bryant et al., 1993; Wolsan, 1999; see also Wyss and Flynn, 1993). Recent molecular analyses, however, (1) ally these taxa with one another in a single monophyletic group, and (2) place this group outside a clade comprised of Mustelidae plus Procyonidae (Ledje and Árnason, 1996a, b; Dragoo and Honeycutt, 1997; Flynn et al., 2000).
METHODS
Data Collection
To permit further resolution of previously enigmatic or controversial relationships, we more than doubled the sampling of caniform species beyond those in our previous analyses (from 17 to 42 species; including 38 arctoids and 4 canids, representing all traditional caniform “families” and more lower level taxa for controversial groups) and increased both taxonomic and nucleotide coverage for the Feliformia (34 species sampled across all of the major feliform clades) (Appendix 1). This study combined sequence data (6243 base pairs [bp]; all confirmed by double stranded sequencing) from six genes: the mitochondrial protein-coding genes cytochrome b (CYTB) and NADH dehydrogenase subunit 2 (ND2), mitochondrial small subunit 12 ribosomal RNA (12S rRNA), the protein-coding nuclear thyroxine-binding globulin (TBG), and interphotoreceptor retinoid binding protein (IRBP) genes, and the first intron of the nuclear transthyretin (TR-i-I) gene (Table 1).
TABLE 1. Carnivoran gene sequence data.
| Genome . | Gene . | Abbreviation . | No. of bp . |
|---|---|---|---|
| Mitochondrial | Cytochrome b | CYTB | 1149 |
| Small subunit 12 ribosomal RNA | 12S | 1067 | |
| NADH dehydrogenase subunit II | ND2 | 1050 | |
| Nuclear | Transthyretin intron 1‡ | TR-i-1 | 1491 |
| Interphotoreceptor retinoid binding protein | IRBP | 1043 | |
| Thyroxine-binding globulin | TBG | 443 |
| Genome . | Gene . | Abbreviation . | No. of bp . |
|---|---|---|---|
| Mitochondrial | Cytochrome b | CYTB | 1149 |
| Small subunit 12 ribosomal RNA | 12S | 1067 | |
| NADH dehydrogenase subunit II | ND2 | 1050 | |
| Nuclear | Transthyretin intron 1‡ | TR-i-1 | 1491 |
| Interphotoreceptor retinoid binding protein | IRBP | 1043 | |
| Thyroxine-binding globulin | TBG | 443 |
‡Includes a 384-bp CanSINE present only in the Caniformia.
| Genome . | Gene . | Abbreviation . | No. of bp . |
|---|---|---|---|
| Mitochondrial | Cytochrome b | CYTB | 1149 |
| Small subunit 12 ribosomal RNA | 12S | 1067 | |
| NADH dehydrogenase subunit II | ND2 | 1050 | |
| Nuclear | Transthyretin intron 1‡ | TR-i-1 | 1491 |
| Interphotoreceptor retinoid binding protein | IRBP | 1043 | |
| Thyroxine-binding globulin | TBG | 443 |
| Genome . | Gene . | Abbreviation . | No. of bp . |
|---|---|---|---|
| Mitochondrial | Cytochrome b | CYTB | 1149 |
| Small subunit 12 ribosomal RNA | 12S | 1067 | |
| NADH dehydrogenase subunit II | ND2 | 1050 | |
| Nuclear | Transthyretin intron 1‡ | TR-i-1 | 1491 |
| Interphotoreceptor retinoid binding protein | IRBP | 1043 | |
| Thyroxine-binding globulin | TBG | 443 |
‡Includes a 384-bp CanSINE present only in the Caniformia.
We concatenated the individual gene sequences into composite data sets for further analysis, as the ability of sequence data to uncover phylogenetic patterns can be augmented through the combination of multiple gene sequences into longer concatenated sequences (Teeling et al., 2000). Prerequisites for combination (concatenation) of sequence data have been extensively debated in the literature, however. Some authors have argued for a “total evidence” approach, whereby all available data are included in an analysis (e.g., Eernisse and Kluge, 1993; Allard and Carpenter, 1996; Nixon and Carpenter, 1996). Considerable empirical work has demonstrated that total evidence data combination can be superior to congruence-based approaches and that the effects of partition incongruence are minimal on phylogenetic analyses (e.g., Allard and Carpenter, 1996; Baker and DeSalle, 1997; Baker et al., 2001; Flynn and Nedbal, 1998). However, we noted in the course of this analysis that ambiguity in the results (especially under maximum parsimony) tended to be concentrated around those taxa that were less thoroughly sampled across the set of gene sequences. As such, we investigated the potential effect of incompleteness (in terms of missing gene sequences) on the precision of phylogenetic analysis.
All of the phylogenetic analyses performed produced unrooted phylogenetic networks of the carnivoran ingroup taxa. As the divergence between Carnivora and other clades of extant eutherian mammals likely occurred prior to 60 to 65 Mya and differentiation within crown-clade Carnivora all occurred more recently than 50 Mya (Flynn and Wesley-Hunt, 2005), the inclusion of distantly related outgroups likely would introduce long evolutionary branches and large amounts of homoplasy to a phylogenetic analysis. These effects could degrade the power of the analysis to resolve the ambiguous interrelationships among major carnivoran clades. To test this, several preliminary analyses were performed using various outgroups; all of these replicated a monophyletic Carnivora, with a basal split separating monophyletic Feliformia and Caniformia clades. Carnivoran monophyly and this bifurcation at the base of the Carnivora have been previously well established, based on both molecular and morphologic data (e.g., Flynn and Galiano, 1982; Flynn et al., 1988, 2000; Wyss and Flynn, 1993; Flynn and Nedbal, 1998; Yoder et al., 2003). We performed extensive maximum parsimony analyses, including assessments of robustness of support for internal nodes (bootstrap proportions and decay indices) and complementary Bayesian phylogenetic analyses.
Nucleotide sequence data were newly sequenced for this study or compiled from previously published studies and GenBank accessions by the authors and others. Sequencing methods for new sequence data for TR-i-1, ND2, CYTB, and IRBP are similar to those described in previous analyses (Dragoo and Honeycutt, 1997; Flynn and Nedbal, 1998; Flynn et al., 2000). Polymerase chain reaction (PCR) for new TBG data was performed using 5′-TGCCACTCTCTACAAGATG-3′ forward primer and 5′-GTGTTTGGCTTGAGGTCTT-3′ reverse primer. The following parameters were employed: initial denaturation at 95°C for 1 min, annealing at 95°C → 54°C → 72°C 1 min each for 30 cycles, and a final extension at 72°C for 3 min.
Sequence data were joined to existing alignments (Árnason et al., 1995; Dragoo and Honeycutt, 1997; Yoder et al., 2003) by eye. In general, we advocate an approach in which unavailable sequence data are coded as missing data in the assembled data matrix. This avoids potential problems caused by the creation of chimeric taxa when taxa assumed to represent monophyletic groups are spliced together (see Malia et al., 2003). However, in one case, sequence data from two very closely related species were used as a single taxon (Mydaus javanensis and Mydausmarchei), assuming that the variation between these particular congeners was insignificant compared to that among the higher level study taxa. The concatenated nucleotide sequence data from six genes comprised a total of 6243 aligned bp for 76 carnivoran taxa (42 caniform taxa and 34 feliform taxa). Carnivoran gene sequence data used in this study is detailed in Table 1. Sources of published data and new sequence data (GenBank accession numbers AY750579 to AY750681) are listed in Appendix 1.
Phylogenetic Analyses
Phylogenetic analyses were performed on the 76-taxon carnivoran ingroup, as well as both the 42-taxon caniform and 34-taxon feliform clade subsets, using the 6243 bp of sequence data detailed above (see Fig. 1, Fig. 2, Fig. 3 and Fig. 4, illustrated summary of major groups shown in Fig. 5). As the monophyly of the Carnivora and its two major subclades (Feliformia, Caniformia) is well documented, all analyses were performed using unrooted networks of the ingroup taxa to avoid introduction of spurious homoplasy due to extremely long branches and distant relationships to even the nearest eutherian outgroup. Preliminary analyses were performed using several different sets of outgroup taxa (e.g., Manis, Elephas, Loxodonta, Equus, Bos, Sus, Homo). The results of these pilot analyses confirm that addition of outgroups does not create topologies within the Carnivora that are incongruent with those obtained when using only an unrooted network of the ingroup taxa, although character optimizations and nodal support values obviously would change, depending on the specific outgroups included. As such, we root the carnivoran phylogenies for analysis assuming only the following constraints: monophyly of Carnivora (in all-Carnivora analyses) and monophyly of the Feliformia and Caniformia (in analyses of each of these subclades). Phylogenetic analyses were performed using both maximum parsimony and Bayesian inference as optimization criteria.
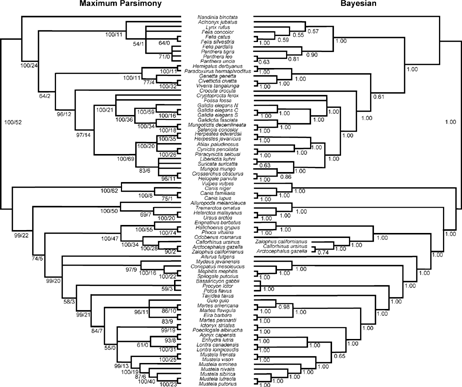
FIGURE 1. Results of the all-Carnivora analyses. Left: Majority-rule consensus of maximum parsimony bootstrap analysis; 1000 heuristic search replicates, with 10 random sequence additions per replicate. Nodal numbers are BP/DI. Right: Majority-rule consensus cladograms of 1.5 million generations of the MCMC analysis of the Bayesian phylogenetic analysis, discarding 100,000 generations as burn-in. Nodal numbers are posterior probabilities.
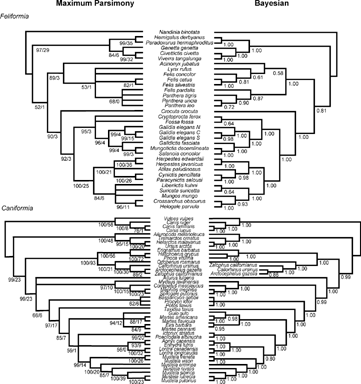
FIGURE 2. Results of the two analyses of feliform-only (top) and caniform-only (bottom) subclades of the Carnivora. Left: Majority-rule consensus of maximum parsimony bootstrap analysis; 1000 heuristic search replicates, with 10 random sequence additions per replicate. Nodal numbers are BP/DI. Right: Majority-rule consensus cladograms of 1.5 million generations of the MCMC analysis of the Bayesian phylogenetic analysis, discarding 100,000 generations as burn-in. Nodal numbers are posterior probabilities.
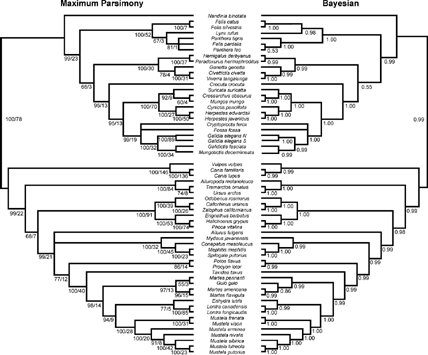
FIGURE 3. Results of the all-Carnivora analyses using a reduced-taxa ingroup, restricted to only those taxa that were sampled for three or more sequences. Left: Majority-rule consensus of maximum parsimony bootstrap analysis; 1000 heuristic search replicates, with 10 random sequence additions per replicate. Nodal numbers are BP/DI. Right: Majority-rule consensus cladograms of 1.5 million generations of the MCMC analysis of the Bayesian phylogenetic analysis, discarding 100,000 generations as burn-in. Nodal numbers are posterior probabilities.
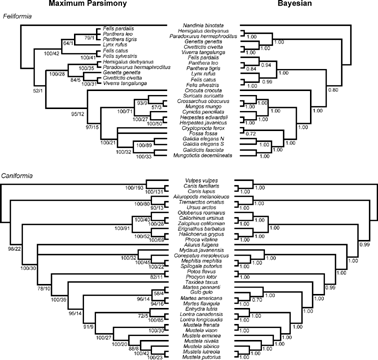
FIGURE 4. Results of the two analyses of feliform (top) and caniform (bottom) subclades of the Carnivora using a reduced-taxa ingroup restricted to only those taxa that were sampled for three or more sequences. Left: Majority-rule consensus of maximum parsimony bootstrap analysis; 1000 heuristic search replicates, with 10 random sequence additions per replicate. Nodal numbers are BP/DI. Right: Majority-rule consensus cladograms of 1.5 million generations of the MCMC analysis of the Bayesian phylogenetic analysis, discarding 100,000 generations as burn-in. Nodal numbers are posterior probabilities.
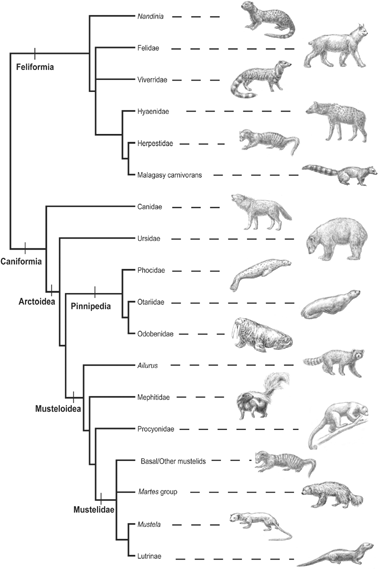
FIGURE 5. A schematic cladogram representing the major evolutionary relationships recovered in this analysis of Carnivora. Illustrations of representative taxa for major lineages include (from top): Nandinia binotata; Felidae (Lynxrufus); Viverridae (Viverra zibetha); Hyaenidae (Crocutacrocuta); Herpestidae (Mungos mungo); Malagasy carnivorans (Eupleres goudotii); Canidae (Canislupus); Ursidae (Ursusamericanus); Phocidae (Phocavitulina); Otariidae (Zalophus californianus); Odobenidae (Odobenus rosmarus); Ailurus fulgens; Mephitidae (Mephitismephitis); Procyonidae (Potosflavus); Mustelidae, basal/other mustelids (generalized schematic representing diverse taxa [African polecat and striped marten, badger, etc.]); Mustelidae, Martes-group (Gulo gulo); Mustelidae, Mustela (Mustelafrenata); Mustelidae, Lutrinae (Lontra canadensis).
Maximum Parsimony
Maximum parsimony (MP) analyses were performed with PAUP** (version 4.10b; Swofford, 2002) using heuristic searches, performed with ten random sequence addition iterations, holding one tree for each iteration, and the tree bisection-reconnection algorithm. Separate analyses were conducted on three partitions of the ingroup taxa: all-Carnivora, Feliformia-only, and Caniformia-only. Robustness of support for the internal nodes was evaluated using bootstrap proportions (BPs) and Bremer decay indices (DIs). Bootstrap proportions (Felsenstein, 1985) for the MP analyses of the concatenated molecular data sets were obtained using heuristic searches for 1000 bootstrap replicates, with 10 random sequence additions per replicate, holding one tree for each iteration. Decay indices represent the number of extra steps required for a clade not to be unequivocally supported (Bremer, 1988). Bremer decay indices were calculated for the strict consensus of the most parsimonious cladograms (100 random sequence additions, heuistic search) using AutoDecay (v 5.0; Eriksson, 2001), and are presented for those nodes that were resolved in the majority-rule consensus of the bootstrap analyses. Figure 1, Figure 2, Figure 3 and Figure 4 (left) present the majority-rule consensus for each maximum parsimony bootstrap analysis with nodal numbers representing BP/DI.
Bayesian Inference
To complement the parsimony analyses, the phylogenetic relationships for each of the taxa sets analyzed under MP (caniform and feliform subsets, all-carnivoran taxa) were also estimated with Bayesian techniques (Rannala and Yang, 1996; Yang and Rannala, 1997; Larget and Simon, 1999), using MrBayes (Version 3.0b4, Huelsenbeck and Ronquist, 2001). Bayesian analysis provides a powerful tool for reconstruction phylogenetic relationships, allowing efficient searching of parameter spaces for complex likelihood models, which can be applied over different partitions of a data set (Huelsenbeck and Ronquist, 2001).
Likelihood ratio tests were employed to determine the substitution model for the Bayesian analysis, using the program MODELTEST (version 3.06; Posada and Crandall, 1998). We employed a six-parameter time-reversible likelihood model, with correction for variation in rates by site and for invariant sites (GTR+Γ+I). However, the sequence data in this study comprise potentially fundamentally different classes of data (mitochondrial protein-coding genes, nuclear protein-coding gene, nuclear intron sequences, and a mitochondrial small subunit rRNA sequence). As such, model parameters should not be assumed to be identical across all data partitions, and were allowed to vary, both among sequences (e.g., TR-i-I versus IRBP) and among codon positions (e.g., 1st position versus 3rd position within CYTB). The unlinking of parameter values across gene partitions and codon positions allows different classes of data, which are likely under different selective constraints, to be modeled with separate parameter values that are optimized for the observed data in each of the a priori–determined classes. The model of evolution might differ among genes or classes of data, and therefore global use of GTR+Γ+I may overparameterize the model in some cases. However, several recent studies demonstrate that model underparameterization is likely to lead to incorrect phylogenetic reconstructions (topologies), and that model overparameterization is less likely to do so (Erixon et al., 2003; Lemon and Moriarity, 2004). We therefore were conservative and applied the more complicated model to capture the most accurate topology.
Analyses were run using the Metropolis-coupled Markov chain, Monte Carlo algorithm in MrBayes, with flat priors. Four separate chains were run for 1,500,000 generations, swapping chains every 5th generation and sampling trees every 100th generation. Inspection of plots of the lnL for the “cold” chain suggests convergence was reached by approximately the 50,000th generation in each analysis. To ensure trees were sampled after convergence, we discarded the first 100,000 generations as burn-in. Posterior probabilities for topologies were then assessed as the proportion of the trees sampled after burn-in in which that particular topology was observed. Figures 1, 2, 3, 4 (right) present the majority-rule consensus cladograms of each Bayesian phylogenetic analysis with nodal numbers representing the posterior probabilities.
Missing Data
As stated above, when sequence data were not available for a taxon, the characters in the data matrix were coded as missing data. Missing data in phylogenetic reconstruction is often considered to be an obstacle for the accurate reconstruction of evolutionary relationships. Although this problem frequently is associated with morphological (especially paleontological) analyses (see discussion in Kearney and Clark, 2003), use of incompletely sampled concatenated sequence data, such as in this study, can present a situation analogous to osteological characters coded for a poorly preserved fossil taxon. In effect, the lack of a gene sequence (e.g., IRBP) for a given taxon can be considered comparable to non-preservation of an anatomical element (e.g., the basicranium). To examine the potential effects of missing data on the accuracy and precision of our phylogenetic analyses, the above procedures were repeated for the feliform-only, caniform-only, and all-carnivoran taxa partitions separately, incorporating a restricted set of better-sampled ingroup taxa (i.e., only those that were represented in the data matrix by at least three sequences).
RESULTS
A noteworthy aspect of the phylogenetic analyses is the high degree of congruence among the recovered topologies for the three ingroup sets and between the two optimization methods (parsimony and Bayesian inference). In general, the phylogenetic relationships within the Carnivora were well resolved in the analyses incorporating the entire carnivoran ingroup (Fig. 1), and the same topologies were also were recovered in the analyses of the more restricted taxon subsets (Fig. 2). In most cases, those nodes that are unambiguously resolved also have robust support. For example, in the all-carnivoran analyses (Fig. 1), most resolved nodes have parsimony BP > 70% and DI = 5–74 (except for two terminal clades linked at BP/DI = 75/1 [Canis lupus/C. familiaris] and 71/0 [Felis pardalis + Panthera spp.]) and Bayesian posterior probabilities = 0.91–1.00. Less than 15% of the nodes have BP = 50–69%/DI = 0–3 or posterior probabilities between 0.56 and 0.82. Although all of the analyses performed using an all-carnivoran ingroup were treated as unrooted networks, every result was consistent with the aforementioned assumption of a bifurcation between monophyletic caniform and feliform lineages at the base of Carnivora. We discuss specific phylogenetic results for Caniformia and Feliformia separately below.
Caniformia
Ledje and Árnason (1996a, 1996b; using CYTB and 12S), Flynn and Nedbal (1998; using morphology, Tr-i-I, CYTB, 12S), and Flynn et al. (2000; using Tr-i-I, CYTB, 12S, 16S) all identified four primary monophyletic clades within the extant Caniformia: Canidae, Ursidae, Pinnipedia, and Musteloidea (sensu lato [s.l.], including Ailurus, mephitids, procyonids and mustelids). In Ledje and Árnason's (1996a, 1996b) studies, the monophyly of each clade was highly supported under bootstrap resampling; however, no strong support could be recovered for the relative interrelationships among these lineages, as their data were unable to resolve a basal polytomy among the four major caniform clades plus the red panda (Ailurus). Flynn and Nedbal (1998) and Flynn et al. (2000) found strong support for monophyly of additional clades within the Caniformia, including the Arctoidea (Ursidae + Pinnipedia + Musteloidea [s.l.]), as well as an otariid-odobenid subclade within Pinnipedia. The results of this study agree with those findings and place Canidae as the sister taxon to the remainder of the caniform clades (Arctoidea).
Within Arctoidea, Flynn and Nedbal (1998) and Flynn et al. (2000) found support for a procyonid-mustelid clade, joined in an unresolved polytomy with mephitids and Ailurus, but were not able to unambiguously resolve the position of this musteloid group relative to Pinnipedia and Ursidae. The analyses in the current study resolve Ursidae as the sister group to all other arctoid taxa, which itself comprises two major clades: (1) the Pinnipedia, including the Phocidae (“true” seals), Otariidae (sea lions) and Odobenidae (walrus), and (2) its sister taxon the Musteloidea (s.l.). In all of our analyses, pinniped monophyly is unambiguous (Lento et al., 1995; Ledje and Árnason, 1996a, 1996b; Flynn and Nedbal, 1998; Flynn et al., 2000; herein). Within the pinnipeds there is a basal split between the Phocidae and an Odobenus-Otariidae clade (Ledje and Árnason, 1996a, 1996b; Flynn and Nedbal, 1998). Although otariids are monophyletic with respect to the walrus, the hypothesized interrelationships of otariid taxa vary between the parsimony and Bayesian analyses. MP analyses support a closer relationship between Zalophus and Arctocephalus. This is consistent with the findings of Wynen et al. (2001), based on partial CYTB and mitochondrial control region sequences, in which the fur seals (Callorhinus and Arctocephalus) do not form a monophyletic group with respect to the sea lions (Zalophus). In contrast, both Bayesian analyses (all-carnivoran and caniform-only) recover a closer relationship between Callorhinus and Arctocephalus (Figs. 1 and 2), supporting fur seal monophyly within the Otariidae. The parsimony support for a sister-taxon relationship between Zalophus and Arctocephalus appears to be strong, whereas the Bayesian posterior probabilities for the alternative topology are not significant. The apparent conflict between the two methods (albeit only weak conflict in the Bayesian analyses) may be due to poor gene coverage of Arctocephalus, for which only two of the six genes used in this study were sampled.
The position of the pinniped clade within Arctoidea is somewhat more complicated. All Bayesian analyses clearly support the placement of ursids and pinnipeds as successive sister groups to the Musteloidea (s.l.) (P = 1.0 and 0.99, for the all-carnivoran and caniform-only analyses, respectively; Figs. 1 and 2). Bininda-Emonds et al. (1999) found weak support for this topology in a supertree analysis of previously published phylogenies (but see critique of this method in Flynn et al., 2000), whereas Flynn and Nedbal (1998) found conflicting support between MP and maximum likelihood analyses of primary character data. Flynn and Nedbal's (1998) analyses combined morphologic data and several of the sequences used in the current study and recovered weak support for the placement of the pinnipeds as the most basal group in the arctoids, with Ursidae as the sister-group to the Musteloidea (s.l.) under maximum parsimony. Their maximum likelihood analyses of only molecular data resolved arctoid interrelationships in the same manner as presented here.
In contrast to the unambiguous results of the Bayesian approaches, MP analysis of all-carnivoran taxa also supports this position for the pinnipeds, but only moderately robustly (BP 74%; Fig. 1). Strikingly, if only the caniform taxa are considered in a maximum parsimony analysis, there is extremely strong support for this topology (BP 99%; Fig. 2), mirroring all the Bayesian results. It is important to note that this single node is the only one for which parsimony BP values are significantly altered between the analysis of the restricted caniform-only ingroup and the all-carnivoran analysis (Figs. 1 and 2). We have investigated possible explanations for this phenomenon, discussed further below.
The Musteloidea (s.l.) as a whole remains a well-supported clade (as in previous studies, e.g., Flynn and Nedbal, 1998; Flynn et al., 2000). However, the polytomy at the base of this clade, among skunks (mephitids), Ailurus (red panda), and the procyonid-mustelid clade (Flynn et al., 2000) cannot be definitively resolved. Examination of bipartition frequencies among parsimony bootstrap replicates indicates that the most frequent association for Ailurus is basal to other musteloids at 47% and with the clade of Mydaus + skunks (Flynn et al., 2000) at 27%. No other position for Ailurus is supported above 5% in the bootstrap. In contrast, Bayesian analyses resolve this basal musteloid polytomy, definitively placing the red panda as the sister taxon to all other musteloids. In none of the analyses (MP or Bayesian) was Ailurus closely allied to either Ursidae (Vrana et al., 1994) or Procyonidae (O'Brien et al., 1985; Slattery and O'Brien, 1995). Traditionally the skunks have been classified within the Mustelidae as a subfamily (Mephitinae), more closely allied to the other mustelids (weasels, polecats, otters, etc.) than any are to the Procyonidae (Bryant et al., 1993; Wolsan, 1999; see also Wyss and Flynn, 1993; Bininda-Emonds et al., 1999). Recent phylogenetic analyses, using various combinations of mitochondrial and nuclear sequences, have recovered a monophyletic clade of skunks that is distinct from a more restricted mustelid clade (e.g., Vrana et al., 1994; Ledje and Árnason, 1996b; Dragoo and Honeycutt, 1997; Flynn et al., 2000). In addition, the stink badgers are also conventionally placed in the Mustelidae, within the Melinae subfamily of Old World badgers (Wozencraft, 1993), although several recent molecular phylogenies instead allied them with skunks (Dragoo and Honeycutt, 1997; Flynn et al., 2000). The results of the Bayesian analyses in the current study strongly support the monophyly of the skunks in a distinct clade (Mephitidae), and therefore the paraphyly of the traditionally conceived Mustelidae (as the mephitids are not the nearest relatives of other mustelids, but rather form the sister-group to a mustelid [s.s.] + procyonid clade). These analyses also clearly document that the stink badgers (represented here by Mydaus) are more closely related to the skunks than to other arctoids (as in the morphological phylogeny of Bryant et al., 1993). Although the monophyly of the skunk + stink badger clade is also strongly supported by parsimony, the BP support for the position of this clade relative to mustelids (s.s.) and procyonids is relatively weak (BP 58% and 66% for all-carnivoran and caniform-only analyses, respectively; Figs. 1 and 2). The BP values for the monophyly of the Procyonidae are also low (BP 59% and 62%, respectively), and neither parsimony nor Bayesian analyses are able to resolve the interrelationships of the three procyonid taxa. As with the otariid situation, this lack of resolution also is likely due to poor sampling for Bassaricyon (see below). This study is unable to resolve a position for Old World badgers, as unfortunately none were sampled for this analysis . The position of this group will need to be assessed further to determine whether they indeed are monophyletic, and, if so, if they will remain closely allied to mustelines and lutrines, or rather are they more closely related to stink badgers (as traditionally portrayed, and therefore also to the skunks). Two recent molecular phylogenies (Koepfli and Wayne, 2003; Sato et al., 2004) found some support for placing several meline taxa (e.g., Meles meles, Melogale moschata, Arctonyx collaris) within the Mustelidae, indicating that they may be polyphyletic with respect to other mustelids (Melogale potentially lying within a clade containing lutrines and a paraphyletic “Mustelinae,” and Meles in a basal position with respect to other mustelid [s.s.] taxa). It should be noted, however, that topologies and support varied in different analyses, and mephitids and Ailurus were not included in those analyses.
Within the Mustelidae (s.s), we find topologies similar to those recovered in other recent phylogenetic analyses. Bayesian analyses significantly resolved all of the internal nodes, with the exception of the weak support for interrelationships among the mustelines (weasels), the lutrines (otters), and a clade uniting two African mustelids (Ictonyx and Poecilogale) (BP < 50%, P = 0.65; Fig. 1). Parsimony, however, did not strongly resolve many of the internal relationships within this clade (Fig. 1).
In all analyses, the genus Martes was paraphyletic, consistent with the earlier findings of Stone and Cook (2002; using complete CYTB and partial nuclear aldolase C gene sequences). In our MP analysis, the wolverine (Gulo) is situated in a basal polytomy with various species of Martes, and Eira is grouped with Martes pennanti alone. Bayesian analysis further refines this topology, finding robust support for a basal split between an Eira + M. pennanti clade and a clade allying the wolverine (Gulo) with M. flavigula and M. americana (Figs. 1 and 2). Stone and Cook (2002) found support for an alternative topology, uniting M. pennanti with Gulo to the exclusion of other mustelids. Sato et al. (2003; using CYTB and IRPB) found weak support for a Gulo +M. flavigula clade and consistently found Gulo to be more closely related to M. flavigula and M. americana, as found in this study. Sato et al. (2003) did not, however, include either Eira or M. pennanti in their analysis. Recently, Koepfli and Wayne (2003; combined data from CYTB and five nuclear STS markers) recovered a topology essentially identical to the one presented here, although they noted that the strength of support for uniting Eira with M. pennanti was weak and only supported by CYTB.
Lutrine monophyly was questioned by Koepfli and Wayne (1998), based on their MP analyses of CYTB sequence data. They found that, under a wide range of parsimony weighting schemes, otters were paraphyletic with respect to mustelines (weasels). However, their maximum likelihood analyses also tended to recover a monophyletic Lutrinae (Koepfli and Wayne, 1998: their fig. 4). Later these authors found stronger support for a monophyletic lutrine clade, when incorporating nuclear sequence data in the phylogenetic analyses (Koepfli and Wayne, 2003). In the current study, the relationship of the lutrines to other musteline taxa is ambiguous under MP (weak support for lutrine monophyly, BP 61%/DI = 0; Fig. 1), although a demonstrably paraphyletic Lutrinae also was not recovered. In contrast, our Bayesian analyses (P = 1.0; Fig. 1) unanimously support the monophyly of the otters (Lutrinae). Three of the taxa in our study (Aonyx, Poecilogale, Ictonyx) have poor gene coverage, however, and much of the ambiguity in the parsimony results relating to whether otters are monophyletic or nonmonophyletic and to the weak support for the interrelationships among mustelids (s.s.) in general may simply be the result of missing sequence data. This will be explored further below. As in the Koepfli and Wayne (1998; 2003) studies, all mustelines are more closely related to the otters than to any other musteloids, although their position relative to various “melines” remains uncertain (as none were sampled in our study).
Feliformia
In general, the results of our analyses for feliform carnivorans conform to earlier studies of this group. In all analyses, there is strong support placing Nandinia (African palm civet) as the sister taxon to all other extant feliform taxa (Fig. 1), confirming earlier demonstrations that Viverridae, as traditionally defined, is paraphyletic (implicit in Hunt, 1987, 1989; Veron, 1995; Flynn and Nedbal, 1998; Yoder et al., 2003; Gaubert and Veron, 2003). Additionally, all of Madagascar's endemic carnivoran taxa form a monophyletic clade, indicating that this radiation resulted from a single dispersal event (Yoder et al., 2003; Yoder and Flynn, 2003). This clade forms the sister-group to the Herpestidae (s.s.), and Hyaenidae (represented in this analysis by Crocuta) is the sister-group to the herpestid + Malagasy carnivorans clade (Flynn and Nedbal, 1998; Yoder et al., 2003; Yoder and Flynn, 2003; Gaubert and Veron, 2003). Lastly, the species of Felidae and a restricted Viverridae s.s. (excluding Nandinia, those Malagasy carnivorans traditionally classified as “viverrids,” and possibly also the Asiatic linsangs [Prionodon]) each form strongly supported monophyletic groups (Fig. 1).
In contrast to the clear resolution of the basal radiations within the caniform lineage, the relative positions among the major groups of Feliformia, which individually are strongly supported as monophyletic (Felidae, Viverridae [s.s.], and clade of Hyaenidae and Herpestidae + Malagasy carnivorans), remain ambiguously resolved by the molecular data (see Flynn et al., 1988, for comparable ambiguity suggested by morphological data). Both MP and Bayesian analyses of all-carnivoran taxa place the Viverridae as the sister group to the (Hyaenidae + (Herpestidae + Malagasy carnivoran)) clade, and thus reconstruct the Felidae, then Nandinia, as the sequential outgroups to this clade (see: Flynn and Nedbal, 1998; Yoder et al., 2003; Gaubert and Veron, 2003). However, the support for this topology is weak (BP = 64%, DI = 2, P = 0.61; Fig. 1). When the analysis is repeated using the feliform-only taxa subset, the relative positions of Felidae and Viverridae (s.s) are reversed, with Felidae most closely allied with the (Hyaenidae + (Herpestidae + Malagasy carnivoran)) clade, although these results are also unpersuasive (BP = 52%, DI = 1, P = 0.81; Fig. 2).
Flynn and Nedbal (1998), Yoder et al. (2003), Yoder and Flynn (2003), and Gaubert and Veron (2003) all recovered Viverridae as the sister-group to the combined (Hyaenidae + (Herpestidae + Malagasy carnivoran)) clade. In Flynn and Nedbal (1998; Tr-i-I only) and Gaubert and Veron (2003; Tr-i-I and CYTB only) that topology is weakly supported (parsimony BP = 61%, DI = 1; ML BP = 78%, insignificant branch length in Flynn and Nebal [1998]; < 70%, not measured, < 70%, and significant branch length for the same measures in Gaubert and Veron [2003], respectively). This topology in Yoder et al. (2003) had weak parsimony support (BP = 61%), but strong posterior support in their Bayesian analysis (P = .99). However, all of these prior studies incorporated only Pantheraleo and Felissilvestris as exemplar felids, and employed many fewer genes than in the current analysis. The addition of more taxa within the Felidae (and throughout the feliforms, and caniform outgroups, in general) and the augmentation of the gene sequence data in this analysis serve to highlight the ambiguity of the evolutionary relationships at this node. The inability of these more extensive analyses to conclusively resolve these basal relationships suggests the possibility that there may have been an early and rapid radiation within this group between the three primary feliform lineages: (1) Felidae (possibly also including Prionodon [Gaubert and Veron, 2003]), (2) the restricted Viverridae, and (3) the lineage (Hyaenidae + (Herpestidae + Malagasy carnivoran)).
Interrelationships within the Felidae are little resolved. In both MP and Bayesian analyses, Felis catus (the domestic cat) and F. silvestris (the wild cat) are grouped as sister taxa, although the strength of support for this node is somewhat weak under parsimony in the all-carnivoran analysis (Fig. 1). When only the feliform taxa are analyzed, the strength of support for this node improves under parsimony (Fig. 2). Beyond this, strong support for the internal relationships within Felidae does not exist. Weak support for a clade uniting Panthera leo, P. tigris, and P. uncia appears in the Bayesian analyses, along with weak support for allying Felis pardalis (ocelot) with these taxa. Unfortunately, these analyses are unable to robustly resolve relationships within this group.
It is possible that this is due in some part to ambiguity introduced by missing data, and perhaps by the proportionally lower taxon sampling of felids (relative to herpestids and Malagasy carnivorans). All of the felid taxa in this study are represented by at least half of the potential sequences (Appendix 1), with three notable exceptions: the puma (Felisconcolor), the snow leopard (Panthera uncia), and the cheetah (Acinonyx jubatus), each of which is known only for a single gene. This is important, as the position of Acinonyx appears to be incompatible between our MP and Bayesian results (although with only weak support for the conflicting placements in each; Figs. 1 and 2). Similarly, in the Bayesian analyses P. uncia is placed as the closest relative of the lion, to the exclusion of the tiger, which is surprising given traditional taxonomic and phylogenetic interpretations (Figs. 1 and 2). Analyses of the data sets restricted to those taxa with three or more sequences better resolve some of the internal relationships of the Felidae. However, the ambiguous relationships among these taxa, especially that of the snow leopard, should be better resolved in the future with increased sequence data and more comprehensive sampling of felid taxa.
Within the other major feliform lineages (i.e., Viverridae s.s., Herpestidae, Malagasy carnivorans), the taxon interrelationships are congruent with earlier studies based on less complete gene and taxon sampling (e.g., Flynn and Nedbal, 1998; Yoder et al., 2003; Yoder and Flynn, 2003; Gaubert and Veron, 2003) for species in common among these studies. The current study enhances the previous work by providing more comprehensive gene and feliform taxon sampling, greater resolution of interrelationships in particular parts of the tree, or more robust support for various nodes within these more terminal feliform clades.
DISCUSSION
The Effect of Missing Data
Several recent studies have suggested that increasing the number of sampled taxa can enhance the accuracy of phylogenetic analyses (e.g., Rannala et al., 1998; Zwickl and Hillis, 2002), and this argument was a primary motivation behind the increased taxonomic sampling effort in the present data matrix. Although more intensive sampling has led to the resolution of previously ambiguous relationships, in our analyses of both the entire ingroup (all-Carnivora) and each of the more restricted taxa subsets, we noted ambiguous nodes and a reduction of resolution and strength of support (especially with regard to BP values in the MP analyses) for the relationships of several taxa. Most of these were centered on species for which sampling was poor across the set of gene sequences analyzed in this study. For example, in the analyses of the caniform taxa, relationships involving the Procyonidae were poorly constrained: (1) monophyly of Procyonidae itself was not strongly supported; (2) the three taxa remained in a polytomy within the Procyonidae; and (3) the position of Procyonidae relative to the Mustelidae [s.s.] and the clade comprised of skunks and Mydaus (Mephitidae) was unresolved (Figs. 1 and 2). However, one of the three taxa, Bassaricyon, was represented by only two gene sequences (Appendix 1). The poor resolution of both the internal relationships among these particular taxa and among the more inclusive clades to which they belong led us to investigate the possibility that poorly sampled taxa (those that did not have some sequence data for at least three of the six genes) were contributing to the ambiguity in the results of the phylogenetic analyses, particularly under the bootstrap resampling technique used to assess support in MP.
We therefore constructed restricted taxa sets by pruning from the data set those taxa that did not possess at least three partial sequences (all partial sequences include at least half of the relevant gene/sequence). This resulted in the removal of eight caniform taxa: Aonyx, Arctocephalus, Bassaricyon, Canisrufus, Eira, Ictonyx, Helarctos, Poecilogale, and nine feliform taxa: Acinonyx, Atilax, Felisconcolor, Galidia elegans (central population), Helogale, Liberiictis, Pantherauncia, Paracynictis, Salanoia. The maximum parsimony and Bayesian analyses were repeated using these restricted taxa sets, following the methods detailed above. Results of the restricted taxa analyses are presented for the entire carnivoran ingroup (Fig. 3), and for separate caniform and feliform taxa subsets (Fig. 4).
Caniformia
The large-scale evolutionary relationships recovered for the caniform carnivorans using the reduced-taxa sets are identical to those presented above, including a basal split between the Canidae and the Arctoidea, placement of the Ursidae as the sister-group to all other arctoid taxa, and monophyly of the musteloid and pinniped clades (compare Figs. 1 and 3). The reduced-taxa analyses for the Caniformia further resolved several of the ambiguous relationships in the all-taxa analyses. Consideration of only those taxa with three or more sequences resolved two nodes that previously were not well supported under all-taxa maximum parsimony analyses (Fig. 4). The phylogenetic position of the Mephitidae as the sister clade to a Procyonidae + Mustelidae (s.s) clade is strongly supported in both the MP and Bayesian analyses for the reduced-taxa sets (Fig. 4). The phylogenetic position of the clade comprised of Martes, Eira, and Gulo was only weakly supported under parsimony for the all-taxa analyses. In contrast, MP analyses for the reduced-taxa sets increase the strength of support for this node, thereby mirroring the results of the Bayesian all-taxa analyses (Fig. 4).
Lutrine monophyly was questioned by Koepfli and Wayne (1998), and the results of the initial all-taxa MP analyses herein failed to recover strong support for a monophyletic clade of otters (although Bayesian analyses unambiguously supported otter monophyly). However, upon removal of the short-clawed otter (Aonyx), which is sampled only for CYTB and a partial ND2 sequence, otters were recovered as a monophyletic group (Lutrinae) with improved support under MP. Similarly, paraphyly of traditional “Mustelinae” was only weakly indicated in all-taxa analyses but is robustly supported in both all-carnivoran and caniform-only reduced-taxa analyses, with the monophyletic clade of Mustela species more closely related to Lutrinae than to other taxa in the “Martes group,” which are traditionally placed within the Mustelinae (Figs. 3 and 4; see also Koepfli and Wayne, 2003).
Two nodes that were not resolvable in the original analyses also remain ambiguous in the MP analyses of the reduced-data sets. First, the phylogenetic position of the red panda remains in a polytomy with the mephitids and the clade uniting procyonids + mustelids (s.s.), in both the all-carnivoran and caniform-only analyses (Fig. 4). Bayesian analyses, however, continue to place Ailurus as the sister taxon to all other musteloids. Second, the reduced-taxa analyses yield the same pattern of resolution for the pinniped node in the all-taxa analyses. When Caniformia is considered alone, the phylogenetic position of the pinniped clade is strongly supported as the sister group to the Musteloidea (Fig. 4). However, inclusion of a feliform outgroup in the all-carnivoran analysis reduces the support for this topology, even when only well-sampled taxa are incorporated (Fig. 3).
As in all the original analyses, Martes paraphyly was recovered in every analysis of reduced-taxa sets. However, the phylogenetic relationships of Martes and other mustelines, recovered upon the exclusion of Eira, highlights one of the few areas where MP and Bayesian tree topologies are in conflict with one another. That Gulo is allied with species of Martes, and that M. flavigula and M. americana are sister taxa to the exclusion of other taxa in the “Martes group” is unequivocal (Figs. 3 and 4). However, upon removal of the poorly sampled mustelid taxa (including Eira), parsimony recovers Gulo as more closely related to M. pennanti, as also was found by Stone and Cook (2002). In contrast, the Bayesian analyses consistently reconstruct Gulo as the sister taxon to M. flavigula and M. americana (Figs. 3 and 4), as was recovered by Bayesian inference in each of the original analyses (Figs. 1 and 2), as well as by Sato et al. (2003) and Koepfli and Wayne (2003; Gulo more closely related to M. americana: M. flavigula was not sampled in that study). Support for either of these conflicting statements is weak for reduced-taxa analyses of both the entire carnivoran ingroup (BP = 55% versus P = 0.86; Fig. 3) or if only the caniform taxa are considered (BP = 58% versus P = 0.70; Fig. 4). This pattern probably represents some ambiguity in the correct placement of Gulo within this subclade of mustelid carnivorans, due to conflicting support from various genes. For each optimization criterion, the reduced-taxa analyses are consistent with the results of the more inclusive analyses, but upon excluding Eira from the mustelid ingroup, the strength of support for allying Gulo with M. flavigula and M. americana drops. Thus, at least in this specific case, the addition of an extra taxon, regardless of its poor gene coverage, adds some information that improves our ability to recover phylogenetic relationships among close relatives in these more terminal branches.
Feliformia
The topologies recovered for feliforms in the analyses of the reduced-taxa sets are generally consistent with the results when the larger suite of ingroup taxa was considered. As with the case of the Caniformia, removal of poorly sampled taxa improves the ability to resolve some of the finer-scale relationships among terminal taxa. For example, both the all-taxa and reduced-taxa analyses document monophyly of the “social mongooses” (Crossarchus, Helogale, Liberiictis, Mungos, Suricata) and imply a single origin of eusociality within the Herpestidae (Austin, 1998; Yoder et al., 2003; Veron et al., 2004). Among this clade of “social mongooses,” the removal of Liberiictis and Helogale in the reduced-taxa analysis demonstrates that Crossarchus and Mungos share a closer evolutionary relationship to each other than either does with Suricata (compare Figs. 1, 2 with 3, 4; see also Yoder et al., 2003 and Veron et al., 2004). This same relationship was recovered but not strongly supported in the Bayesian analyses of the more inclusive taxon data sets, and the previous MP analyses left the relationship of these three social mongoose taxa unresolved in the majority-rule consensus (Figs. 1 and 2).
Interrelationships of taxa within the Viverridae (s.s.) and the Malagasy carnivoran clade are consistent with other analyses (e.g., Yoder et al., 2003; Yoder and Flynn, 2003; Gaubert and Veron, 2003), with generally high nodal support. This is not unexpected, in part because our taxon and gene sampling for these clades is very similar to those in our earlier collaborative studies on the relationships of Malagasy carnivorans (Yoder et al., 2003; Yoder and Flynn, 2003). In the present study, however, we include more genes and many more felids and caniforms than in those studies, and our species sample differs substantially from the viverrid taxa addressed by Gaubert and Veron (2003). It is noteworthy that the increased taxon sampling across both feliforms and caniforms generally reduces parsimony bootstrap support for more terminal nodes, likely due to additional ingroup homoplasy arising from incorporation of more distantly related outgroups, although support for some nodes does increase with improved ingroup sampling and/or gene coverage. Although further emphasizing the monophyly of various subclades within major feliform lineages, including some within the Malagasy carnivoran clade, increased gene and taxon sampling across the Feliformia is still unable to resolve the basal polytomy between Cryptoprocta, Fossa, and the remaining members of this clade.
Recently, Gaubert and Veron (2003) suggested that Prionodon pardicolor (a taxon not sampled in the current study, but placed within the Viverrinae [Viverridae] in recent classifications) is most closely related to Felidae. However, this alternative placement within Feliformia was variably supported (ML branch length significant, but ML bootstrap only 77% and MP bootstrap < 70%) and was based on fewer genes (Tr-i-I and CYTB only) and a much different and smaller sample of taxa (five caniform outgroups and 22 feliform species; mostly viverrines [three species of Viverra, six species of Genetta], with only two felids, two herpsetids, and one species each of Hyaenidae and Malagasy carnivoran). If this placement of Prionodon is corroborated by future analyses, it would further accentuate the non-monophyly of taxa traditionally placed in the Viverridae.
Although we note a general trend toward increased ability to resolve phylogenetic relationships of taxa in this study when highly incomplete taxa are excluded, the missing data problem cannot be used as a universal explanation for observed ambiguities. The removal of three poorly sampled taxa within the Felidae (Felisconcolor, Patherauncia, and Acinonyxjubatus) provided better insight into the internal relationships within this clade. The tiger and lion are grouped in a clade together with the ocelot (Felis pardalis) in both the Bayesian and MP analyses with strong support (Fig. 3). There is strong support for the clade of “small-bodied” felids (the domestic cat F. catus, the wildcat F. silvestris, and the bobcat Lynx rufus) in the Bayesian analyses of reduced taxa sets (P = 0.98 for the all-carnivoran analysis, P = 0.99 for the feliform only analysis; Figs. 3 and 4). Not surprisingly, within this clade, F. catus and F. silvestris are closely related (BP = 100%, P = 1.0 in all analyses). Although the support for a clade of small-bodied felids appears robust in the Bayesian analyses, parsimony analyses unite the bobcat with the tiger + lion + ocelot clade, albeit with only very weak support (BP = 67%, DI = 3; Fig. 3; and BP = 64%, DI = 1; Fig. 4). Therefore, although some of the lack of resolution that we observed in the original analyses may indeed be the result of missing data for three of the felid taxa, there remains a high degree of ambiguity in this family even after their removal. The interrelationships of the Felidae present a difficult phylogenetic problem that was only compounded by effects of missing data in the all-carnivoran analysis.
The greatest ambiguity in the phylogenetic analyses of the Feliformia still rests in the inability of any method to satisfactorily resolve the early split among the three major feliform lineages (Viverridae [s.s], Felidae, and Hyaenidae + Herpestidae + Malagasy carnivorans), following the basal divergence of this entire clade from Nandinia (found in all analyses in this study). As with the original analyses, each of these three clades was recovered as a strongly supported, monophyletic group in both reduced-taxa analyses of all-carnivoran taxa (Fig. 3) and feliforms-only (Fig. 4). However, the removal of poorly sampled taxa did nothing to improve the resolution of the interrelationships among these lineages. As with the results for the entire ingroup, when considering the all-carnivoran reduced-taxa subset, felids are placed as basal to the viverrids (s.s.), although the support for this arrangement is weak (BP = 66%, DI = 3, P = 0.55; Fig. 3). In contrast, when only the Feliformia are considered, MP and Bayesian analyses are at odds over the position of the Felidae and Viverridae (s.s) relative to the Hyaenidae + (Herpestidae + Malagasy carnivoran) clade. For the feliform-only subset, viverrids (s.s) are reconstructed as the sister group to the felids and the hyaenid + (herpestid + Malagasy carnivoran) clade in the Bayesian analyses (Fig. 4), consistent with the feliform-only analysis using all of the ingroup taxa (Fig. 2), although this topology remains weakly supported (P = 0.80). However, parsimony reconstructs Felidae as basal to a combined viverrid + hyaenid + (herpestid + Malagasy carnivoran) clade (Fig. 4), consistent with the all-carnivoran analyses using the entire ingroup (Fig. 1), although again very weakly supported (BP = 52%, DI = 1). Thus, the evolutionary relationships among these three lineages must be considered ambiguous, and poor sampling among taxa in this clade is not the cause of all of the ambiguity in the interrelationships among these three lineages. Currently, the best representation of the evolutionary relationships of these lineages is an unresolved, tripartite split near the base of the extant feliform radiation.
Although the likelihood functions employed by our Bayesian analyses generally were better able to reconstruct phylogenetic relationships when incompletely sampled taxa were included in the analysis, as indicated by an increased ability to resolve polytomies over the parsimony analyses, we note that these analyses were not immune to the effects of missing data. For example, the relationships within the mustelid clade were unresolved by both MP and Bayesian techniques (Figs. 1 and 2), until Aonyx, Ictonyx, and Poecilogale were removed from the analysis (reduced-taxa analyses; Figs. 3 and 4).
This issue of incompletely sampled taxa is analogous to problems often faced by systematists using paleontological data in cladistic analyses, where missing morphology can contribute large numbers of blank cells in the character by taxon matrix. Missing data do not contribute to support for any topological statement in parsimony analyses. Therefore, taxa represented by large numbers of blank cells will tend to be highly mobile, as missing data allow multiple topologies to be consistent with the observed data (Wilkinson, 1994, 1995, 1996).
Wiens (2003a, 2003b), however, has shown through simulation studies that the detrimental effect of missing data on phylogenetic precision is more likely the result of too few characters (in absolute numbers) rather than the proportion of missing charters for any given taxon. Even extremely incomplete data matrices (one half of the taxa coded for only 10% of the characters) can yield correct and completely resolved maximum parsimony phylogenies if the absolute number of characters is very large (i.e., 2000 characters; Wiens 2003b). This makes intuitive sense, as it is the absolute number of unique synapomorphies that serves to support clades under parsimony, and increasing the number of characters therefore increases the probability of sampling at least some synapomorphies that will resolve polytomies. Thus, Wiens (2003b) argued that the proportion of coded characters may not be a good criterion for exclusion of a taxon from an analysis. Indeed, several authors argue that fragmentary taxa are not necessarily an impediment to the accuracy and precision of phylogenetic analyses (Wilkinson, 1995; Nixon and Carpenter, 1996; Kearney, 2002) and should not be excluded a priori.
In the current study the problem is not simply confined to blank cells, which are randomly distributed through a data matrix, as may be the case in a paleontological study. Rather, the incompleteness here is a function of unsampled genes, and missing data between highly incomplete taxa results in cases where there may be little or no overlapping sequence data. Thus, taxa that are poorly sampled not only have fewer total characters from which to sample synapomorphies but also may not have any basis for direct comparison with certain other taxa. For example, within the Felidae Panthera uncia is sampled only for the ND2 gene and has no overlapping sequences in our data matrix with P. leo, Acinonyx jubatus, Felis pardalis, or F. concolor. Nonoverlapping character data can lead to inflation in the number of most parsimonious reconstructions in phylogenetic analyses, and Wilkinson (1995) has advocated a set of rules for safe removal or combination of taxa from a data set, under the concept of taxonomic equivalence. However, reduction of two or more terminal taxa into a single OTU (Operational Taxonomic Unit) makes an assumption that they collectively represent a monophyletic unit with respect to the rest of the ingroup. This can be justified in cases where character data overlap between terminal taxa. Malia (2003) has shown that the assumption of monophyly, and subsequent combination of terminal taxa for nonoverlapping sequence data, can alter the topology of a phylogenetic analysis, not merely degrade its precision.
The results of the current study suggest that simply adding taxa to an analysis will not necessarily aid in resolving ambiguities in phylogenetic relationships. Adding a taxon with large amounts of missing data (i.e., taxa sampled for only a single gene) can reduce the resolution for nodes near that taxon, without aiding in resolving or supporting other more distant nodes. It is important to note that incomplete sampling was not investigated in the previous studies of increased taxon sampling (e.g., Rannala et al., 1998; Zwickl and Hillis, 2002). It is likely that the reduction in the resolution of phylogenetic analyses observed here arises from nonoverlap of gene sequence data among the less well-sampled taxa. This is a concern for comparable supermatrix studies (e.g., Madsen et al., 2001; Murphy et al. 2001a, 2001b), in which gene sequences are concatenated in an effort to increase the resolving power of the analysis, but in which the individual concatenated data sets are not reciprocally complete for the entire set of ingroup taxa. We note that this poses a particular problem for resampling methods such as the bootstrap. As the level of incompleteness of a taxon increases, the possibility that random resampling will draw heavily from characters with unknown states in one or more taxa increases as well. This contributes to a higher number of most parsimonious reconstructions and lower resolution within and among bootstrap replicates, lowering bootstrap support as taxa for which entire genes are absent are added.
The Effect of Additional IRBP Data
As we were completing our original set of phylogenetic analyses, Sato et al. (2003) published a study of mustelid phylogenetic relationships, based on CYTB and IRBP sequence data. The comparable topologies of the Sato et al. (2003) phylogeny were consistent with what we already had recovered. However, although we incorporate more gene sequences in the present analysis and our taxonomic sampling is larger, our IRBP sequence data was more limited for our sample of caniform taxa than for feliform taxa. Prior to the publication of the Sato et al. (2003) study, we only had IRBP data for nine caniform species (21%), compared to 21 feliforms (60%). We decided to add the IRBP data from Sato et al. (2003) to our existing matrix and reanalyze the entire data set to evaluate the potential effects of these missing data. IRBP data for 10 overlapping taxa from Sato et al. (2003) were incorporated into our data matrix (Mustela ermina, M. lutreola, M. nivalis, M. putorius, M. sibirica, M. vision, Martes americana, M. flavigula, Gulo gulo, Enhydra lutris), doubling the coverage of IRBP in our caniform data set. We repeated MP and Bayesian analyses on both the all-carnivoran and caniform-only taxa sets, following the same methodologies, to determine if integration of additional IRBP data would alter the results of our analysis, either in the topology recovered or in the strength of support for various caniform clades. It was not necessary to replicate the feliform-only analyses as data were only added for caniform taxa.
All recovered topologies were unaffected by the addition of the Sato et al. (2003) IRBP sequence data. The additional data for the 10 mustelid species did not alter the pattern of, or affect support for, hypotheses of interrelationships of either these specific taxa or other musteloids under either MP or Bayesian analysis. These results are summarized for the all-carnivoran taxa set in Table 2.
TABLE 2. Comparison of support for selected nodes before and after integration of additional IRBP. Data from Sato et al. (2003) in all-Carnivora analyses.
| . | Original analysis . | With added IRBP data . | ||
|---|---|---|---|---|
| Node . | BP . | Bayesian posterior probability . | BP . | Bayesian posterior probability . |
| Felidae monophyletic | 100 | 1.00 | 99 | 1.00 |
| Viverridae (s.s.) monophyletic | 100 | 1.00 | 100 | 1.00 |
| Herpestidae (s.s.) monophyletic | 100 | 1.00 | 100 | 1.00 |
| Malagasy Carnivora monophyletic | 100 | 1.00 | 100 | 1.00 |
| Canidae monophyletic | 100 | 1.00 | 100 | 1.00 |
| Arctoidea monophyletic | 99 | 1.00 | 99 | 1.00 |
| Ursidae monophyletic | 100 | 1.00 | 100 | 1.00 |
| Pinnipedia monophyletic | 100 | 1.00 | 100 | 1.00 |
| Pinnipedia + Musteloidea monophyletic | 74 | 1.00 | 61 | 1.00 |
| Musteloidea monophyletic | 99 | 1.00 | 97 | 1.00 |
| Mephitidae monophyletic | 97 | 1.00 | 97 | 1.00 |
| Procyonidae + Mustelidae (s.s.) | 58 | 1.00 | 58 | 1.00 |
| Mustelidae (s.s.) monophyletic | 99 | 1.00 | 99 | 1.00 |
| Eira + Martes pennanti | 83 | 1.00 | 85 | 1.00 |
| Gulo + M. flavigula and M. americana | < 50 | 0.98 | < 50 | 1.00 |
| Mustela + Lutrinae | < 50 | 0.65 | < 50 | 0.93 |
| Lutrinae monophyletic | 61 | 1.00 | 60 | 1.00 |
| . | Original analysis . | With added IRBP data . | ||
|---|---|---|---|---|
| Node . | BP . | Bayesian posterior probability . | BP . | Bayesian posterior probability . |
| Felidae monophyletic | 100 | 1.00 | 99 | 1.00 |
| Viverridae (s.s.) monophyletic | 100 | 1.00 | 100 | 1.00 |
| Herpestidae (s.s.) monophyletic | 100 | 1.00 | 100 | 1.00 |
| Malagasy Carnivora monophyletic | 100 | 1.00 | 100 | 1.00 |
| Canidae monophyletic | 100 | 1.00 | 100 | 1.00 |
| Arctoidea monophyletic | 99 | 1.00 | 99 | 1.00 |
| Ursidae monophyletic | 100 | 1.00 | 100 | 1.00 |
| Pinnipedia monophyletic | 100 | 1.00 | 100 | 1.00 |
| Pinnipedia + Musteloidea monophyletic | 74 | 1.00 | 61 | 1.00 |
| Musteloidea monophyletic | 99 | 1.00 | 97 | 1.00 |
| Mephitidae monophyletic | 97 | 1.00 | 97 | 1.00 |
| Procyonidae + Mustelidae (s.s.) | 58 | 1.00 | 58 | 1.00 |
| Mustelidae (s.s.) monophyletic | 99 | 1.00 | 99 | 1.00 |
| Eira + Martes pennanti | 83 | 1.00 | 85 | 1.00 |
| Gulo + M. flavigula and M. americana | < 50 | 0.98 | < 50 | 1.00 |
| Mustela + Lutrinae | < 50 | 0.65 | < 50 | 0.93 |
| Lutrinae monophyletic | 61 | 1.00 | 60 | 1.00 |
| . | Original analysis . | With added IRBP data . | ||
|---|---|---|---|---|
| Node . | BP . | Bayesian posterior probability . | BP . | Bayesian posterior probability . |
| Felidae monophyletic | 100 | 1.00 | 99 | 1.00 |
| Viverridae (s.s.) monophyletic | 100 | 1.00 | 100 | 1.00 |
| Herpestidae (s.s.) monophyletic | 100 | 1.00 | 100 | 1.00 |
| Malagasy Carnivora monophyletic | 100 | 1.00 | 100 | 1.00 |
| Canidae monophyletic | 100 | 1.00 | 100 | 1.00 |
| Arctoidea monophyletic | 99 | 1.00 | 99 | 1.00 |
| Ursidae monophyletic | 100 | 1.00 | 100 | 1.00 |
| Pinnipedia monophyletic | 100 | 1.00 | 100 | 1.00 |
| Pinnipedia + Musteloidea monophyletic | 74 | 1.00 | 61 | 1.00 |
| Musteloidea monophyletic | 99 | 1.00 | 97 | 1.00 |
| Mephitidae monophyletic | 97 | 1.00 | 97 | 1.00 |
| Procyonidae + Mustelidae (s.s.) | 58 | 1.00 | 58 | 1.00 |
| Mustelidae (s.s.) monophyletic | 99 | 1.00 | 99 | 1.00 |
| Eira + Martes pennanti | 83 | 1.00 | 85 | 1.00 |
| Gulo + M. flavigula and M. americana | < 50 | 0.98 | < 50 | 1.00 |
| Mustela + Lutrinae | < 50 | 0.65 | < 50 | 0.93 |
| Lutrinae monophyletic | 61 | 1.00 | 60 | 1.00 |
| . | Original analysis . | With added IRBP data . | ||
|---|---|---|---|---|
| Node . | BP . | Bayesian posterior probability . | BP . | Bayesian posterior probability . |
| Felidae monophyletic | 100 | 1.00 | 99 | 1.00 |
| Viverridae (s.s.) monophyletic | 100 | 1.00 | 100 | 1.00 |
| Herpestidae (s.s.) monophyletic | 100 | 1.00 | 100 | 1.00 |
| Malagasy Carnivora monophyletic | 100 | 1.00 | 100 | 1.00 |
| Canidae monophyletic | 100 | 1.00 | 100 | 1.00 |
| Arctoidea monophyletic | 99 | 1.00 | 99 | 1.00 |
| Ursidae monophyletic | 100 | 1.00 | 100 | 1.00 |
| Pinnipedia monophyletic | 100 | 1.00 | 100 | 1.00 |
| Pinnipedia + Musteloidea monophyletic | 74 | 1.00 | 61 | 1.00 |
| Musteloidea monophyletic | 99 | 1.00 | 97 | 1.00 |
| Mephitidae monophyletic | 97 | 1.00 | 97 | 1.00 |
| Procyonidae + Mustelidae (s.s.) | 58 | 1.00 | 58 | 1.00 |
| Mustelidae (s.s.) monophyletic | 99 | 1.00 | 99 | 1.00 |
| Eira + Martes pennanti | 83 | 1.00 | 85 | 1.00 |
| Gulo + M. flavigula and M. americana | < 50 | 0.98 | < 50 | 1.00 |
| Mustela + Lutrinae | < 50 | 0.65 | < 50 | 0.93 |
| Lutrinae monophyletic | 61 | 1.00 | 60 | 1.00 |
Variable Support for the Position of Pinnipedia under MP Analyses of Different Taxon Samples
In the original MP analyses, BP support for the phylogenetic position of the Pinnipedia decreased markedly in the all-carnivoran analysis relative to the caniform-only analysis (Figs. 1 and 2, respectively). The caniform-only analyses provide strongly supported resolution of pinniped relationships within Arctoidea, indicating that they are most closely related to the Musteloidea, with ursids as the sister-group to all other extant arctoids (BP 99% and DI = 23, P = 0.99; Fig. 2). The phylogeny for the all-carnivoran data set shows substantially lower BP and DI values for this node (BP 74% and DI = 5; Fig. 1), although the Bayesian support remained unequivocal (P = 1.00). This pattern of strong support for a basal position of Ursidae within Arctoidea when only the caniform taxa were analyzed, and the reduction of support when the entire carnivoran ingroup was analyzed was observed for both the original analyses (compare Figs. 1 and 2) and the reduced-taxa analyses (compare Figs. 3 to 4). Thus, this pattern cannot be explained as ambiguity introduced by poor gene sampling and missing data.
The problem reduces to the relative positions of the three major arctoid lineages (Ursidae, Pinnipedia, and Musteloidea), all of which are unambiguously monophyletic in all analyses. Examination of the alternative bipartitions in the bootstrap reconstruct pinnipeds with Ursidae in 16.2% of the bootstrap replicates, whereas Ursidae is linked to Musteloidea in 8.4% of the replicates. No other topology that includes the Pinnipedia was recovered in more than 5% of the replicates. Thus, upon inclusion of feliform taxa in the analysis, the pinniped clade is drawn to the ursids in some bootstrap resamplings. It is important to note that the node separating monophyletic Musteloidea from ursids and pinnipeds is not affected regardless of the configuration of the three lineages, including Ailurus.
This behavior has some of the characteristics of long-branch attraction, although the erosion of support for a single specific internal branch is unusual. The incorporation of the feliform taxa creates a more inclusive clade (Carnivora), with sequence data for many taxa beyond the more restricted clade to which the pinnipeds belong, in what amounts to adding a feliform “outgroup” to Caniformia. In this more inclusive all-carnivoran analysis, the dramatic changes in support are observed only for this particular node within the caniform “ingroup.” Including the feliform taxa yields a substantial erosion of bootstrap support for the pinniped-musteloid linkage, whereas all other nodes remain virtually unchanged (contrast Fig. 2 and Fig. 1 BP and DI values). In addition, there are no further changes in the topology between these two analyses, and there is no corresponding loss of support in the complementary Bayesian analysis of all-carnivoran taxa (Fig. 1: P = 1.0; Fig. 2: P = 0.99). In our analyses, the erosion of support for this particular caniform node is caused by the addition of a relatively distant “outgroup,” the feliform taxa. A similar phenomenon was observed by Lento et al. (1995), who noticed that adding taxa to the data set could erode the strength of support of previously well-supported nodes while leaving the strength of support at neighboring, well-supported nodes unchanged.
To better understand the cause of the decreased support for this node, outgroup analyses and maximum parsimony bootstrap analyses were performed using 17 selected pairs of feliform species. To avoid potential confounding effects of poor gene sampling, all of the taxa used as outgroups were represented by a majority of the sequences in this study. The results show that the support for the position of the pinnipeds can vary dramatically when specific species are used, and therefore also when these species are included as part of the broader feliform outgroup taxon sample in the all-carnivoran analysis. The resulting BP supporting the node uniting pinnipeds + musteloids varies between 55% and 97%, depending on the specific pair of feliform species selected as exemplar taxa to represent Feliformia (Table 3). In some cases, the nodal support remains virtually unchanged from the extremely high levels observed in the caniform-only analyses; for example, when various combinations of felids and/or hyaenids are used as the feliform outgroup. In contrast, the support levels are extremely low (range = 55–70%; mean = 62%) when any of the pairs of herpestid species from the clade of social mongooses (Crossarchus, Suricata, Mungos) are used as the outgroup. Each of the BP values for the three possible pairings of these taxa is lower than when any other pairing of feliform taxa is used (range = 74–97%; mean = 86%). Bootstrap support is also higher when the three social mongoose taxa are excluded entirely from outgroup pairs (range = 79–97%; mean = 87.5%). Inclusion of these specific taxa, therefore, greatly influences the reduction in support for the single node defining Pinnipedia + Musteloidea in the all-carnivoran analyses, although their influence is most marked in isolation.
TABLE 3. Bootstrap support for the “pinniped node” using selected herpestid taxa as outgroup to the Caniformia.
| Outgroup pair . | BP . | Mean . |
|---|---|---|
| Crossarchus, Suricata | 55 | — |
| Crossarchus, Mungos | 70 | — |
| Suricata, Mungos | 61 | — |
| All others pairings | 74–97 | 86 |
| All pairings completely excluding social mongooses | 79–97 | 87.5 |
| Outgroup pair . | BP . | Mean . |
|---|---|---|
| Crossarchus, Suricata | 55 | — |
| Crossarchus, Mungos | 70 | — |
| Suricata, Mungos | 61 | — |
| All others pairings | 74–97 | 86 |
| All pairings completely excluding social mongooses | 79–97 | 87.5 |
| Outgroup pair . | BP . | Mean . |
|---|---|---|
| Crossarchus, Suricata | 55 | — |
| Crossarchus, Mungos | 70 | — |
| Suricata, Mungos | 61 | — |
| All others pairings | 74–97 | 86 |
| All pairings completely excluding social mongooses | 79–97 | 87.5 |
| Outgroup pair . | BP . | Mean . |
|---|---|---|
| Crossarchus, Suricata | 55 | — |
| Crossarchus, Mungos | 70 | — |
| Suricata, Mungos | 61 | — |
| All others pairings | 74–97 | 86 |
| All pairings completely excluding social mongooses | 79–97 | 87.5 |
Thus, although all the parsimony analyses recover precisely the same topology, each resolving the node defining the position of pinnipeds as the sister group to the musteloids (almost certainly because of the informativeness of the nuclear gene data at this hierarchical level; Table 4), resampling methods (BP) yield greatly reduced support because of the noise introduced by homoplasious similarities (especially in the mitochondrial gene data; Table 4) with various taxa in the feliform outgroup. In a case such as this, the addition of taxa from a fairly distantly related outgroup (the feliform taxa), introducing homoplasy that erodes resampling support for a node that is otherwise well supported when only ingroup taxa are analyzed, is likely responsible for the ambiguity observed in the parsimony bootstrap results. Thus we consider the topology observed in all analyses to accurately represent the phylogenetic relationships among family-level groupings of arctoid carnivorans, with pinnipeds as the sister-group to the musteloids (s.l.).
TABLE 4. Average character retention indices by gene for the three main taxa sets.
| . | Mean character RI . | ||
|---|---|---|---|
| . | All-Carnivora . | Caniformia-Only . | Feliformia-Only . |
| All sequence data | 0.583 | 0.526 | 0.556 |
| nDNA | 0.859 | 0.846 | 0.825 |
| TR-i-I | 0.881 | 0.852 | 0.850 |
| IRBP | 0.713 | 0.590 | 0.796 |
| TBG | 0.843 | 0.899 | 0.690 |
| mtDNA | 0.491 | 0.448 | 0.487 |
| 12S | 0.471 | 0.406 | 0.500 |
| ND2 | 0.516 | 0.468 | 0.510 |
| CYTB | 0.466 | 0.439 | 0.459 |
| . | Mean character RI . | ||
|---|---|---|---|
| . | All-Carnivora . | Caniformia-Only . | Feliformia-Only . |
| All sequence data | 0.583 | 0.526 | 0.556 |
| nDNA | 0.859 | 0.846 | 0.825 |
| TR-i-I | 0.881 | 0.852 | 0.850 |
| IRBP | 0.713 | 0.590 | 0.796 |
| TBG | 0.843 | 0.899 | 0.690 |
| mtDNA | 0.491 | 0.448 | 0.487 |
| 12S | 0.471 | 0.406 | 0.500 |
| ND2 | 0.516 | 0.468 | 0.510 |
| CYTB | 0.466 | 0.439 | 0.459 |
| . | Mean character RI . | ||
|---|---|---|---|
| . | All-Carnivora . | Caniformia-Only . | Feliformia-Only . |
| All sequence data | 0.583 | 0.526 | 0.556 |
| nDNA | 0.859 | 0.846 | 0.825 |
| TR-i-I | 0.881 | 0.852 | 0.850 |
| IRBP | 0.713 | 0.590 | 0.796 |
| TBG | 0.843 | 0.899 | 0.690 |
| mtDNA | 0.491 | 0.448 | 0.487 |
| 12S | 0.471 | 0.406 | 0.500 |
| ND2 | 0.516 | 0.468 | 0.510 |
| CYTB | 0.466 | 0.439 | 0.459 |
| . | Mean character RI . | ||
|---|---|---|---|
| . | All-Carnivora . | Caniformia-Only . | Feliformia-Only . |
| All sequence data | 0.583 | 0.526 | 0.556 |
| nDNA | 0.859 | 0.846 | 0.825 |
| TR-i-I | 0.881 | 0.852 | 0.850 |
| IRBP | 0.713 | 0.590 | 0.796 |
| TBG | 0.843 | 0.899 | 0.690 |
| mtDNA | 0.491 | 0.448 | 0.487 |
| 12S | 0.471 | 0.406 | 0.500 |
| ND2 | 0.516 | 0.468 | 0.510 |
| CYTB | 0.466 | 0.439 | 0.459 |
However, the phenomenon of a degradation in the support for a specific and otherwise robustly supported topology, coupled with our observations of the effect of using small outgroup subsets that include specific problematic taxa (here, the social mongooses), raises significant general cautions about the danger of using small numbers of exemplar taxa as proxies for a diverse outgroup in phylogenetic analyses. It also emphasizes the potential effects on nodal support of including such taxa as part of a broader suite of relatively distantly related outgroup taxa (i.e., feliforms as outgroup to caniforms; lineages that likely diverged at least 50 million years ago; Flynn and Wesley-Hunt, 2005; Wesley-Hunt, and Flynn, 2005) or in a comprehensive analysis (i.e., all-carnivorans, with relationships among the entire suite of study taxa not specified a priori, in an unrooted analysis). Had our analysis of caniform phylogeny been rooted using one of these problematic social mongoose taxa as an exemplar of the Herpestidae, or primarily emphasized parsimony bootstrap support levels in an all-carnivoran analysis, we might have missed strong evidence resolving this previously ambiguous node.
Implications for Reconstructing Ancestral Body Size in Arctoidea
The pattern of caniform interrelationships recovered in the current study suggests a novel inference about the evolution of body size in the arctoids. Among living taxa, ursids and pinnipeds are large-bodied forms relative to arctoid outgroups and most of the extant musteloids (Ailurus, mephitids, mustelids (s.s.), procyonids). As pinnipeds and ursids are now resolved as sequential outgroups to the musteloids, this implies that large body size may be primitive for Arctoidea, with a subsequent size reduction in the Musteloidea. A preliminary investigation into this pattern was undertaken for the caniform clade. Body size data were obtained for 149 extant caniform species from the MOMv3.3 database (Smith et al., 2003) and arranged in rank order from smallest (Mustelanivalis, ∼ 100 g) to largest (Miroungaleonina, ∼ 1600 kg). Each species in the database was assigned to one of the following 10 clades (Fig. 5): Canidae; Urisdae; Pinnipedia; Ailuridae; Mephitidae; Procyonidae; and Mustelidae: (a) basal/“other” taxa (including Taxidea, and taxa such as Meles and Melogale, following Koepfli and Wayne [2003], Sato et al. [2004]; as well as the African polecat taxa Ictonyx and Poecilogale), (b) Martes group (including Eira, Gulo and Martes), (c) Lutrinae, and (d) Mustela. These clades were ranked 1 to 10 according to the branching order of the phylogeny in Figure 1. A highly significant negative correlation was observed when clade rank was compared to rank body size (Spearman rank correlation, (rs)c = −0.68, P ≪ 0.001). However, the assignment of the clades Lutrinae and Mustela group to clade ranks 9 and 10, respectively, in this comparison is arbitrary, as the partition between the two branches is trivial. That is, the two topologies (Martes group, (Lutrinae, Mustela)) and (Martes group, (Mustela, Lutrinae)) are identical. As the lutrines are generally larger than members of the Mustela clade, arbitrarily assigning the later group the higher clade rank could cause the r-value to be artificially high. As such, a second rank correlation was performed, switching the clade ranks for these two groups, and a similarly strong negative correlation was observed ((rs)c = −0.60, P ≪ 0.001).
An inference based only on living taxa must be viewed with caution, however, as incorporation of stem fossil taxa might change the optimization for living clades, as could the phylogenetic positions of entirely extinct lineages (e.g., amphicyonids) within the Caniformia. For example, Wang (1997) proposed that the large-bodied fossil Simocyon is the nearest relative to the ailurines, which could indicate that large rather than small body size is primitive for the Ailurus lineage. This would in turn strengthen the evidence for large body size being primitive for Arctoidea, as all three outgroups to the remaining musteloids would then be large. Fossil stem pinnipedimorphs, such as Enaliarctos and Pinnarctidion (Berta and Sumich, 1999; Deméré et al., 2003), are consistent in body size with the modern members of the clade, although early ursids range in body size from small to large. Incorporation of fossil stem taxa, and robust inferences of their phylogenetic positions relative to living clades, are likely to be essential to an accurate reconstruction of ancestral body size.
CONCLUSIONS
The present phylogenetic analysis of Carnivora represents the most comprehensive sampling of taxa and genes yet undertaken for the group, including 76 carnivoran ingroup taxa (analyzed together [all-Carnivora] as well as in separate caniform and feliform subsets) and a concatenated sequence of 6243 bp from six genes. Maximum parsimony (MP) and Bayesian methods were remarkably congruent for virtually all analyses of each of these sets of carnivoran taxa, in both the topologies recovered and the nodal support measures (MP bootstrap and Bremer decay indices, Bayesian posterior probabilities).
We recovered all previously robustly supported (in analyses using various sets of morphological and molecular data) higher level carnivoran clades, including monophyly of Caniformia, Feliformia, Arctoidea, Pinnipedia, Musteloidea, Procyonidae + Mustelidae (s.s.), and a clade of (Hyaenidae + (Herpestidae + Malagasy carnivorans)). All of the traditional “families,” except Viverridae and Mustelidae, were robustly supported as monophyletic clades.
This study was able to robustly resolve the position of the marine carnivoran clade (Pinnipedia) within arctoids, documenting a sister-group relationship between pinnipeds and musteloids (s.l), with ursids as the sister clade to the pinniped-musteloid clade. However, this robust support, observed in both the Bayesian and MP analyses for the caniform-only analysis, was not recovered in the all-Carnivora parsimony analysis. Notably, this was the only node for which the strength of support was significantly altered. Analysis of selected feliform outgroup taxa reveals that the BP support for this clade also drops significantly when any of the “social mongoose” herpestid taxa are used in the outgroup, implying that the use of exemplar taxa as proxies for entire clades with diverse evolutionary histories should be approached with caution.
This pattern of caniform interrelationships suggests a novel inference about the ancestral body size for arctoids, implying that large body size may be primitive for Arctoidea, with size reduction in the Musteloidea (s.l.). An inference based only on living taxa must be viewed with caution, however. Incorporation of fossil stem taxa, and robust inferences of their phylogenetic positions relative to living clades, might change the optimization for living groups, and thus inclusion of fossils is likely to be essential for accurate reconstructions of ancestral body size.
At lower hierarchical levels within Caniformia we recovered phylogenetic patterns previously observed, but with varying degrees of support, in several other studies. Within Pinnipedia, Phocidae and Otariidae were both monophyletic, Odobenidae was allied more closely with the otariids than the phocids, and otariid interrelationships differ in Bayesian and parsimony reconstructions. The skunks and closely allied stink badgers (Mydaus) form a monophyletic clade (Mephitidae), not nested within the Mustelidae (as in traditional phylogenies), but rather is the nearest outgroup to a clade of Procyonidae + Mustelidae (s.s.). Within Mustelidae s.s., the genus Martes is paraphyletic with respect to both Gulo and Eira, although their interrelationships remain ambiguous. Lutrinae is monophyletic and unambiguously supported in all Bayesian analyses, although MP analyses failed to strongly support a monophyly (but did not support diphyly either); removal of poorly sampled taxa increased MP support for monophyly somewhat.
Topologies of the major feliform lineages conform to those observed in our earlier collaborative studies, based on lesser taxon or gene sampling and/or fewer caniform outgroups. We confirmed that Nandinia is the sister taxon to all other extant feliform carnivorans. The monophyly of each of the other three major feliform clades (Viverridae (s.s.), Felidae, and a clade of Hyaenidae + (Herpestidae + Malagasy carnivorans)) is robust in all analyses, but the relative phylogenetic positions of these three lineages is not resolvable at present. Failure to resolve these interrelationships is not due primarily to sampling issues, but rather is likely the result of a rapid radiation early in the evolutionary history of the extant feliforms. There is little resolution within Felidae, although some support exists for (a) allying the snow leopard (Pantherauncia) with P. leo,P. tigris and the ocelot (Felispardalis), and (b) a clade uniting the domestic and wild cats (F. catus and F. silvestris) with the puma (F. concolor). Each analysis recovers a monophyletic clade of Malagasy carnivorans, closely allied with the Herpestidae (s.s.), and “social mongoose” monophyly suggests a single origin of eusociality within Herpestidae. Removal of poorly sampled taxa yields a closer relationship between Crossarchus and Mungos than either shares with Suricata.
Recent studies suggested that increasing taxon sampling can enhance phylogenetic accuracy and resolution; this was generally supported by our more intensive sampling of Carnivora. Incomplete character sampling was not investigated in prior studies of increased taxon sampling, and simply adding taxa to an analysis will not necessarily aid in resolving phylogenetic ambiguities. Adding a taxon with large amounts of missing character data can reduce the resolution for nodes near that taxon without aiding in resolving or supporting other more distant nodes. In various analyses we noted ambiguous nodes and reduction of resolution and strength of support for the relationships of several taxa, typically those for which gene sampling was less complete. We therefore investigated reduced-taxon data sets to assess whether poorly sampled taxa (with sequence data for less than three of the six genes) generally led to greater phylogenetic ambiguity, particularly under MP bootstrap resampling. The reduced-taxa analyses for both the Caniformia and Feliformia further resolved several of the ambiguous relationships in the all-taxa analyses. Although there is a general trend toward increased ability to resolve phylogenetic relationships when highly incomplete taxa are excluded, the missing data problem cannot be used as a universal explanation for observed ambiguities. Character incompleteness poses a particular problem for resampling methods such as the bootstrap. This contributes to a higher number of most parsimonious reconstructions and lower resolution within and among bootstrap replicates, lowering bootstrap support as taxa for which entire genes are absent are added. Bayesian analysis likelihood functions generally were better able to reconstruct phylogenetic relationships than parsimony analyses when incompletely sampled taxa were included, as evidenced by increased resolution and more robust support for various nodes. These Bayesian analyses were not immune, however, to the effects of missing data. This likely arises from nonoverlap of gene sequence data among less well-sampled taxa, which is a concern for similar studies, in which different gene sequences are concatenated in an effort to increase resolving power.
ACKNOWLEDGMENTS
Sequencing for taxa/genes not previously published was carried out in the Field Museum's Pritzker Laboratory for Molecular Systematics and Evolution (operated with generous support from The Pritzker Foundation). This research was supported by NSF grants to J.J.F. and M.A.N. (DEB-9707225) and to Anne D. Yoder, J.J.F., and M.A.N. (DEB-9807045). We are especially grateful to Anne Yoder for our collaborations on key related aspects of phylogenetic analyses of feliforms, particularly the Malagasy carnivorans. We thank the many sources of tissue samples specified in Dragoo and Honeycutt (1997) and Flynn and Nedbal (1998). For new samples cited in Appendix 1, we would like to thank the following individuals: A. Gardner, G. A. Guliuzza, J. Kerbis, P. de Groot, J. W. Dragoo, L. Frank, L. Heaney, and S. M. Goodman, and the following institutions: University of Alaska, Fairbanks Museum, US Fish and Wildlife Service, Audubon Zoo, Brookfield Zoo, Museum of Southwestern Biology University of New Mexico, Albuquerque, University of California, Berkeley, Field Museum of Natural History, Louisiana State University, Museum of Zoology, Metro Toronto Zoo, National Zoological Park, Washington, DC, University of Toronto, Royal Ontario Museum, Shedd Aquarium, San Diego Zoo, and St. Louis Zoo for gracious loans of tissue and/or DNA samples.
APPENDIX 1.
Taxa and sequences used in the phylogenetic analysis. Family-level classification based on Wozencraft (1993). Blank cells = sequence data currently unavailable.
| Family . | Taxon/species . | Common name . | TR . | 12S . | ND2 . | CYTB . | IRBP . | TBG . | Voucher . |
|---|---|---|---|---|---|---|---|---|---|
| Ailuridae | Ailurus fulgens | Red panda | AF0397396 | Y0851112 | AY750613** | X9491911 | AY750608** | AY750652** | NZP 83-671 |
| Canidae | Canis familiaris | Domestic dog | AY750579** | Y0850712 | NC_0020088 | X9492011 | AY750653** | PL2267 | |
| Canis lupus | Gray wolf | AF0397326 | AY17004421 | AY17010321 | AY17007421 | ||||
| Canis rufus | Red wolf | AY750580** | AY750654** | GAG132 | |||||
| Vulpes vulpes | Red fox | AF0397336 | Y0850712 | AY750614** | X9492911 | AY750655** | PL2023 | ||
| Mustelidae | Aonyx capensis | Short clawed or clawless otter | AY750615** | AF0571182 | PL4663 | ||||
| Conepatus mesoleucus | Hog-nosed skunk | AY750581** | U783265 | AY750616** | AY750656** | NK 43633 | |||
| Eira barbara | Tayra | AY750582** | AY750617** | SDZ 588414 | |||||
| Enhydra lutris | Sea otter | AY750583** | Y0851212 | AY750618** | AF0571209 | AB082978‡17 | AY750657** | SA “Nuka” | |
| Gulo gulo | Wolverine | AY750584** | U783335 | AY750619** | X9492111 | AB082962‡17 | BZ-47-F4 | ||
| Ictonyx striatus | Zorilla (African polecat) | AY750585** | U783345 | BZ-880085 | |||||
| Lontra canadensis | North American river otter | AY750586** | U783355 | AY750620** | AF0571219 | AY750620** | PL 2068 | ||
| Lontra longicaudis | Neotropical river otter | AF0397346 | AY750621** | AF0571239 | ALG-14988 | ||||
| Martes americana | American marten | AY750587** | U783365 | AY750622** | AF0571309 | AB082963‡17 | BZ-47-C8 | ||
| Martes flavigula | Yellow-throated marten | AY750588** | AY750623** | AB01236310 | AB082964‡17 | SDZ-32799 | |||
| Martes pennanti | Fisher | AY750589** | AY750624** | AF0571319 | PL-4273 | ||||
| Mephitis mephitis | Striped skunk | AF3069487 | Y0851712 | AY750625** | X9492711 | AY750609** | AY750659** | NK 43631 | |
| Mustela erminea | Ermine (stoat) | AY750590** | AY750626** | AB02610115 | AB082969‡17 | LJB-2435 | |||
| Mustela frenata | Long-tailed weasel | AF0397356 | U783395 | AY750627** | AF0685474 | AY750660** | PL-4656 | ||
| Mustela lutreola | European mink | AY750591** | AY750628** | AB02610515 | AB082972‡17 | PL-3441 | |||
| Mustela nivalis | European common weasel | AY750592** | Y0851512 | AY750629** | AB02610615 | AB082973‡17 | SDZ5001 | ||
| Mustela putorius | European polecat | AY750593** | Y0851612 | AY750630** | AB02610715 | AB082975‡17 | SDZ-33221 | ||
| Mustela sibirica | Siberian weasel (kolinsky) | AY750594** | AY750631** | AB02610815 | SDZ-32108 | ||||
| Mustela vision | American mink | AY750595** | Y0851412 | AY750632** | AB02610915 | AB082977‡17 | LJB-2124 | ||
| Mydaus spp. | Stink badger | AY750596** | U783425 | AY750633** | JDW | ||||
| Poecilogale albinucha | African striped weasel | AY750597** | AY750634** | JCK2891 | |||||
| Spilogale putorius | Spotted skunk | AF3069497 | U783465 | AY750635** | X9492811 | AY750610** | AY750661** | NK 43632 | |
| Taxidea taxus | American badger | AY750598** | U783475 | AY750636** | AF0571329 | PL2542 | |||
| Odobenidae | Odobenus rosmarus | Walrus | AF0397436 | U783435 | AY750637** | X822992 | AY750662** | BZ910053 | |
| Otariidae | Arctocephalus gazella | Southern fur seal | Y0852612 | X822929 | |||||
| Callorhinus ursinus | Northern fur seal | AF0397446 | U1283013 | AY750638** | AY750663** | AF1421 | |||
| Zalophus californianus | California sea lion | AF0397456 | Y0852512 | AY750639** | X82310 2 | AY750612** | AY750664** | LSUMZ-044 | |
| Phocidae | Erignathus barbatus | Bearded seal | AF0397426 | AY17004721 | AY17010421 | AY17007721 | AY750665** | AF-1417 | |
| Halichoerus grypus | Grey seal | NC_0016021 | NC_0016021 | NC_0016021 | |||||
| Phoca vitulina | Harbor seal | AY750599** | AY750640** | X823062 | AY750666** | SLZ-920124 | |||
| Procyonidae | Bassaricyon gabbii | Olingo | Y0851012 | X9493111 | |||||
| Potos flavus | Kinkajou | AF0397376 | U783445 | AY750641** | L2187622 | AY750611** | AY750667** | ALG-14904 | |
| Procyon lotor | Common raccoon | AF0397366 | Y0851012 | AY17004621 | X9493011 | AY17007621 | AY750668** | PL-2093 | |
| Ursidae | Ailuropoda melanoleuca | Giant panda | AF0397386 | Y0852112 | AY750642** | X9491811 | AY750669** | NZP-92765 | |
| Helarctos malayanus | Malayan sun bear | U1889920 | |||||||
| Tremartos ornatus | Spectacled bear | AF0397406 | L2188322 | AY17004521 | U2355420 | AY17007521 | AY750670** | PL-959 | |
| Ursus arctos | Grizzly bear (brown bear) | AF0397416 | Y0851912 | AY750643** | X823082 | AY750671** | BZ “Doo” | ||
| Felidae | Acinonyx jubatus | Cheetah | AY750600** | AUD-688 | |||||
| Felis catus | Domestic cat | Y0850312 | NC_00170014 | AB004238 | Z1181119 | ||||
| Felis concolor | Puma | U3349518 | |||||||
| Felis pardalis | Ocelot | AY750601** | U783315 | AY750672** | JMC-298 | ||||
| Felis silvestris | Wild cat | AF0397246 | AY17004221 | AY17010221 | AY17007221 | AY750673** | SDZ-32188 | ||
| Lynx rufus | Bobcat | AY750602** | AY750644** | AY750674** | PL-2025 | ||||
| Panthera leo | Lion | AF0397256 | Y0850512 | AF0530523 | AY750675** | SLZ-076005 | |||
| Panthera tigris | Tiger | AY750603** | Y0850412 | AY750645** | X823012 | AY750676** | PL-942 | ||
| Panthera uncia | Snow leopard | AY750646** | PL-2087 | ||||||
| Herpestidae | Atilax paludinosus | Marsh mongoose | AY750604** | AY750647** | ROM-C1A | ||||
| Crossarchus obscurus | Dark mongoose (kusimanse) | AF0397266 | AY17004121 | AY17010121 | AY17007121 | ||||
| Cynictis pencillata | Yellow mongoose | AY17002421 | AY17004921 | AY17010621 | AY17007921 | ||||
| Galidia elegans (central) | Ring-tailed mongoose | AF0397276 | AY750648** | SMG-6499 | |||||
| Galidia elegans (northern) | Ring-tailed mongoose | AY17002121 | AY17003921 | AY17009921 | AY17006921 | ||||
| Galidia elegans (southern) | Ring-tailed mongoose | AY17002021 | AY17003821 | AY17009821 | AY17006821 | ||||
| Galidictis fasciata | Broad-striped mongoose | AY17002221 | AY17004021 | AY17010021 | AY17007021 | AY750677** | SMG-7554 | ||
| Helogale parvula | Dwarf mongoose | AY750605** | AY750649** | SDZ 30590 | |||||
| Herpestes edwardsii | Indian gray mongoose | AY17002521 | AY17005021 | AY17010721 | AY17008021 | ||||
| Herpestes javanicus | Small Indian mongoose | AY17002621 | Y0850612 | AY17005121 | AY17010821 | AY17008121 | |||
| Liberiictis kuhni | Liberian mongoose | AY750650** | MTZ | ||||||
| Mungos mungo | Banded mongoose | AY17001721 | AY17003521 | AY17009521 | AY17006521 | ||||
| Mungotictis decemlineata | Narrow-striped mongoose | AY17001621 | AY17003421 | AY17009421 | AY17006421 | ||||
| Paracynictis selousi | Gray meerkat (selous' mongoose) | AY750606** | AY750651** | MC-95935 | |||||
| Salanoia concolor | Brown mongoose | AY750607** | AY18700721 | FMNH-33946 | |||||
| Suricata suricatta | Meerkat (suricate) | AY17002821 | D2889916 | AY17005421 | AY17011121 | AY17008421 | |||
| Hyaenidae | Crocuta crocuta | Spotted hyena | AF0397286 | AY17005721 | AY17011421 | AY17008721 | AY750678** | LF “Sargent” | |
| Viverridae | Civettictis civetta | African civet | AY17002321 | AY17004821 | AY17010521 | AY17007821 | |||
| Cryptoprocta ferox | Fossa | AY17001821 | AY17003621 | AY17009621 | AY17006621 | ||||
| Fossa fossa | Fanaloka (Malagasy civet) | AY17001921 | AY17003721 | AY17009721 | AY17006721 | AY750679** | SMG-7539 | ||
| Genetta servalina | Servaline genet | AY17002921 | AY17005821 | AY17011521 | AY17008821 | ||||
| Hemigalus derbyanus | Banded palm civet | AY17002721 | AY17005221 | AY17010921 | AY17008221 | ||||
| Nandinia binotata | African palm civet | AF0397296 | AY17005321 | AY17011021 | AY17008321 | AY750680** | JCK-2623 | ||
| Paradoxurus hermaphroditus | Common palm civet | AF0397306 | AY17005621 | AY17011321 | AY17008621 | ||||
| Viverra tangalunga | Malayan civet | AF0397316 | AY17005521 | AY17011221 | AY17008521 | AY750681** | LRH-4121 | ||
| Family . | Taxon/species . | Common name . | TR . | 12S . | ND2 . | CYTB . | IRBP . | TBG . | Voucher . |
|---|---|---|---|---|---|---|---|---|---|
| Ailuridae | Ailurus fulgens | Red panda | AF0397396 | Y0851112 | AY750613** | X9491911 | AY750608** | AY750652** | NZP 83-671 |
| Canidae | Canis familiaris | Domestic dog | AY750579** | Y0850712 | NC_0020088 | X9492011 | AY750653** | PL2267 | |
| Canis lupus | Gray wolf | AF0397326 | AY17004421 | AY17010321 | AY17007421 | ||||
| Canis rufus | Red wolf | AY750580** | AY750654** | GAG132 | |||||
| Vulpes vulpes | Red fox | AF0397336 | Y0850712 | AY750614** | X9492911 | AY750655** | PL2023 | ||
| Mustelidae | Aonyx capensis | Short clawed or clawless otter | AY750615** | AF0571182 | PL4663 | ||||
| Conepatus mesoleucus | Hog-nosed skunk | AY750581** | U783265 | AY750616** | AY750656** | NK 43633 | |||
| Eira barbara | Tayra | AY750582** | AY750617** | SDZ 588414 | |||||
| Enhydra lutris | Sea otter | AY750583** | Y0851212 | AY750618** | AF0571209 | AB082978‡17 | AY750657** | SA “Nuka” | |
| Gulo gulo | Wolverine | AY750584** | U783335 | AY750619** | X9492111 | AB082962‡17 | BZ-47-F4 | ||
| Ictonyx striatus | Zorilla (African polecat) | AY750585** | U783345 | BZ-880085 | |||||
| Lontra canadensis | North American river otter | AY750586** | U783355 | AY750620** | AF0571219 | AY750620** | PL 2068 | ||
| Lontra longicaudis | Neotropical river otter | AF0397346 | AY750621** | AF0571239 | ALG-14988 | ||||
| Martes americana | American marten | AY750587** | U783365 | AY750622** | AF0571309 | AB082963‡17 | BZ-47-C8 | ||
| Martes flavigula | Yellow-throated marten | AY750588** | AY750623** | AB01236310 | AB082964‡17 | SDZ-32799 | |||
| Martes pennanti | Fisher | AY750589** | AY750624** | AF0571319 | PL-4273 | ||||
| Mephitis mephitis | Striped skunk | AF3069487 | Y0851712 | AY750625** | X9492711 | AY750609** | AY750659** | NK 43631 | |
| Mustela erminea | Ermine (stoat) | AY750590** | AY750626** | AB02610115 | AB082969‡17 | LJB-2435 | |||
| Mustela frenata | Long-tailed weasel | AF0397356 | U783395 | AY750627** | AF0685474 | AY750660** | PL-4656 | ||
| Mustela lutreola | European mink | AY750591** | AY750628** | AB02610515 | AB082972‡17 | PL-3441 | |||
| Mustela nivalis | European common weasel | AY750592** | Y0851512 | AY750629** | AB02610615 | AB082973‡17 | SDZ5001 | ||
| Mustela putorius | European polecat | AY750593** | Y0851612 | AY750630** | AB02610715 | AB082975‡17 | SDZ-33221 | ||
| Mustela sibirica | Siberian weasel (kolinsky) | AY750594** | AY750631** | AB02610815 | SDZ-32108 | ||||
| Mustela vision | American mink | AY750595** | Y0851412 | AY750632** | AB02610915 | AB082977‡17 | LJB-2124 | ||
| Mydaus spp. | Stink badger | AY750596** | U783425 | AY750633** | JDW | ||||
| Poecilogale albinucha | African striped weasel | AY750597** | AY750634** | JCK2891 | |||||
| Spilogale putorius | Spotted skunk | AF3069497 | U783465 | AY750635** | X9492811 | AY750610** | AY750661** | NK 43632 | |
| Taxidea taxus | American badger | AY750598** | U783475 | AY750636** | AF0571329 | PL2542 | |||
| Odobenidae | Odobenus rosmarus | Walrus | AF0397436 | U783435 | AY750637** | X822992 | AY750662** | BZ910053 | |
| Otariidae | Arctocephalus gazella | Southern fur seal | Y0852612 | X822929 | |||||
| Callorhinus ursinus | Northern fur seal | AF0397446 | U1283013 | AY750638** | AY750663** | AF1421 | |||
| Zalophus californianus | California sea lion | AF0397456 | Y0852512 | AY750639** | X82310 2 | AY750612** | AY750664** | LSUMZ-044 | |
| Phocidae | Erignathus barbatus | Bearded seal | AF0397426 | AY17004721 | AY17010421 | AY17007721 | AY750665** | AF-1417 | |
| Halichoerus grypus | Grey seal | NC_0016021 | NC_0016021 | NC_0016021 | |||||
| Phoca vitulina | Harbor seal | AY750599** | AY750640** | X823062 | AY750666** | SLZ-920124 | |||
| Procyonidae | Bassaricyon gabbii | Olingo | Y0851012 | X9493111 | |||||
| Potos flavus | Kinkajou | AF0397376 | U783445 | AY750641** | L2187622 | AY750611** | AY750667** | ALG-14904 | |
| Procyon lotor | Common raccoon | AF0397366 | Y0851012 | AY17004621 | X9493011 | AY17007621 | AY750668** | PL-2093 | |
| Ursidae | Ailuropoda melanoleuca | Giant panda | AF0397386 | Y0852112 | AY750642** | X9491811 | AY750669** | NZP-92765 | |
| Helarctos malayanus | Malayan sun bear | U1889920 | |||||||
| Tremartos ornatus | Spectacled bear | AF0397406 | L2188322 | AY17004521 | U2355420 | AY17007521 | AY750670** | PL-959 | |
| Ursus arctos | Grizzly bear (brown bear) | AF0397416 | Y0851912 | AY750643** | X823082 | AY750671** | BZ “Doo” | ||
| Felidae | Acinonyx jubatus | Cheetah | AY750600** | AUD-688 | |||||
| Felis catus | Domestic cat | Y0850312 | NC_00170014 | AB004238 | Z1181119 | ||||
| Felis concolor | Puma | U3349518 | |||||||
| Felis pardalis | Ocelot | AY750601** | U783315 | AY750672** | JMC-298 | ||||
| Felis silvestris | Wild cat | AF0397246 | AY17004221 | AY17010221 | AY17007221 | AY750673** | SDZ-32188 | ||
| Lynx rufus | Bobcat | AY750602** | AY750644** | AY750674** | PL-2025 | ||||
| Panthera leo | Lion | AF0397256 | Y0850512 | AF0530523 | AY750675** | SLZ-076005 | |||
| Panthera tigris | Tiger | AY750603** | Y0850412 | AY750645** | X823012 | AY750676** | PL-942 | ||
| Panthera uncia | Snow leopard | AY750646** | PL-2087 | ||||||
| Herpestidae | Atilax paludinosus | Marsh mongoose | AY750604** | AY750647** | ROM-C1A | ||||
| Crossarchus obscurus | Dark mongoose (kusimanse) | AF0397266 | AY17004121 | AY17010121 | AY17007121 | ||||
| Cynictis pencillata | Yellow mongoose | AY17002421 | AY17004921 | AY17010621 | AY17007921 | ||||
| Galidia elegans (central) | Ring-tailed mongoose | AF0397276 | AY750648** | SMG-6499 | |||||
| Galidia elegans (northern) | Ring-tailed mongoose | AY17002121 | AY17003921 | AY17009921 | AY17006921 | ||||
| Galidia elegans (southern) | Ring-tailed mongoose | AY17002021 | AY17003821 | AY17009821 | AY17006821 | ||||
| Galidictis fasciata | Broad-striped mongoose | AY17002221 | AY17004021 | AY17010021 | AY17007021 | AY750677** | SMG-7554 | ||
| Helogale parvula | Dwarf mongoose | AY750605** | AY750649** | SDZ 30590 | |||||
| Herpestes edwardsii | Indian gray mongoose | AY17002521 | AY17005021 | AY17010721 | AY17008021 | ||||
| Herpestes javanicus | Small Indian mongoose | AY17002621 | Y0850612 | AY17005121 | AY17010821 | AY17008121 | |||
| Liberiictis kuhni | Liberian mongoose | AY750650** | MTZ | ||||||
| Mungos mungo | Banded mongoose | AY17001721 | AY17003521 | AY17009521 | AY17006521 | ||||
| Mungotictis decemlineata | Narrow-striped mongoose | AY17001621 | AY17003421 | AY17009421 | AY17006421 | ||||
| Paracynictis selousi | Gray meerkat (selous' mongoose) | AY750606** | AY750651** | MC-95935 | |||||
| Salanoia concolor | Brown mongoose | AY750607** | AY18700721 | FMNH-33946 | |||||
| Suricata suricatta | Meerkat (suricate) | AY17002821 | D2889916 | AY17005421 | AY17011121 | AY17008421 | |||
| Hyaenidae | Crocuta crocuta | Spotted hyena | AF0397286 | AY17005721 | AY17011421 | AY17008721 | AY750678** | LF “Sargent” | |
| Viverridae | Civettictis civetta | African civet | AY17002321 | AY17004821 | AY17010521 | AY17007821 | |||
| Cryptoprocta ferox | Fossa | AY17001821 | AY17003621 | AY17009621 | AY17006621 | ||||
| Fossa fossa | Fanaloka (Malagasy civet) | AY17001921 | AY17003721 | AY17009721 | AY17006721 | AY750679** | SMG-7539 | ||
| Genetta servalina | Servaline genet | AY17002921 | AY17005821 | AY17011521 | AY17008821 | ||||
| Hemigalus derbyanus | Banded palm civet | AY17002721 | AY17005221 | AY17010921 | AY17008221 | ||||
| Nandinia binotata | African palm civet | AF0397296 | AY17005321 | AY17011021 | AY17008321 | AY750680** | JCK-2623 | ||
| Paradoxurus hermaphroditus | Common palm civet | AF0397306 | AY17005621 | AY17011321 | AY17008621 | ||||
| Viverra tangalunga | Malayan civet | AF0397316 | AY17005521 | AY17011221 | AY17008521 | AY750681** | LRH-4121 | ||
**DNA Sequence new to this study.
‡IRBP sequence data from Sato et al. (2003) added for replicate analyses only (see text).
Vouchers for new sequence data are given with the following abbreviations for institutions: (AF) = University of Alaska, Fairbanks Museum; (ALG) = USFWS; (AUD) = Audubon Zoo; (BZ) = Brookfield Zoo; (JWD, NK) = Museum of Southwestern Biology University of New Mexico, Albuquerque; (LF) = University of California, Berkeley; (JCK LRH SMG PL MC GAG) = Field Museum of Natural History; (LSUMZ, JCS, JMC, LJB) = Louisiana State University, Museum of Zoology; (MTZ) = Metro Toronto Zoo; (NZP) = National Zoological Park, Washington D.C.; (ROM) = University of Toronto, Royal Ontario Museum; (SA) = Shedd Aquarium; (SDZ) = San Diego Zoo; (SLZ) = St. Louis Zoo.
| Family . | Taxon/species . | Common name . | TR . | 12S . | ND2 . | CYTB . | IRBP . | TBG . | Voucher . |
|---|---|---|---|---|---|---|---|---|---|
| Ailuridae | Ailurus fulgens | Red panda | AF0397396 | Y0851112 | AY750613** | X9491911 | AY750608** | AY750652** | NZP 83-671 |
| Canidae | Canis familiaris | Domestic dog | AY750579** | Y0850712 | NC_0020088 | X9492011 | AY750653** | PL2267 | |
| Canis lupus | Gray wolf | AF0397326 | AY17004421 | AY17010321 | AY17007421 | ||||
| Canis rufus | Red wolf | AY750580** | AY750654** | GAG132 | |||||
| Vulpes vulpes | Red fox | AF0397336 | Y0850712 | AY750614** | X9492911 | AY750655** | PL2023 | ||
| Mustelidae | Aonyx capensis | Short clawed or clawless otter | AY750615** | AF0571182 | PL4663 | ||||
| Conepatus mesoleucus | Hog-nosed skunk | AY750581** | U783265 | AY750616** | AY750656** | NK 43633 | |||
| Eira barbara | Tayra | AY750582** | AY750617** | SDZ 588414 | |||||
| Enhydra lutris | Sea otter | AY750583** | Y0851212 | AY750618** | AF0571209 | AB082978‡17 | AY750657** | SA “Nuka” | |
| Gulo gulo | Wolverine | AY750584** | U783335 | AY750619** | X9492111 | AB082962‡17 | BZ-47-F4 | ||
| Ictonyx striatus | Zorilla (African polecat) | AY750585** | U783345 | BZ-880085 | |||||
| Lontra canadensis | North American river otter | AY750586** | U783355 | AY750620** | AF0571219 | AY750620** | PL 2068 | ||
| Lontra longicaudis | Neotropical river otter | AF0397346 | AY750621** | AF0571239 | ALG-14988 | ||||
| Martes americana | American marten | AY750587** | U783365 | AY750622** | AF0571309 | AB082963‡17 | BZ-47-C8 | ||
| Martes flavigula | Yellow-throated marten | AY750588** | AY750623** | AB01236310 | AB082964‡17 | SDZ-32799 | |||
| Martes pennanti | Fisher | AY750589** | AY750624** | AF0571319 | PL-4273 | ||||
| Mephitis mephitis | Striped skunk | AF3069487 | Y0851712 | AY750625** | X9492711 | AY750609** | AY750659** | NK 43631 | |
| Mustela erminea | Ermine (stoat) | AY750590** | AY750626** | AB02610115 | AB082969‡17 | LJB-2435 | |||
| Mustela frenata | Long-tailed weasel | AF0397356 | U783395 | AY750627** | AF0685474 | AY750660** | PL-4656 | ||
| Mustela lutreola | European mink | AY750591** | AY750628** | AB02610515 | AB082972‡17 | PL-3441 | |||
| Mustela nivalis | European common weasel | AY750592** | Y0851512 | AY750629** | AB02610615 | AB082973‡17 | SDZ5001 | ||
| Mustela putorius | European polecat | AY750593** | Y0851612 | AY750630** | AB02610715 | AB082975‡17 | SDZ-33221 | ||
| Mustela sibirica | Siberian weasel (kolinsky) | AY750594** | AY750631** | AB02610815 | SDZ-32108 | ||||
| Mustela vision | American mink | AY750595** | Y0851412 | AY750632** | AB02610915 | AB082977‡17 | LJB-2124 | ||
| Mydaus spp. | Stink badger | AY750596** | U783425 | AY750633** | JDW | ||||
| Poecilogale albinucha | African striped weasel | AY750597** | AY750634** | JCK2891 | |||||
| Spilogale putorius | Spotted skunk | AF3069497 | U783465 | AY750635** | X9492811 | AY750610** | AY750661** | NK 43632 | |
| Taxidea taxus | American badger | AY750598** | U783475 | AY750636** | AF0571329 | PL2542 | |||
| Odobenidae | Odobenus rosmarus | Walrus | AF0397436 | U783435 | AY750637** | X822992 | AY750662** | BZ910053 | |
| Otariidae | Arctocephalus gazella | Southern fur seal | Y0852612 | X822929 | |||||
| Callorhinus ursinus | Northern fur seal | AF0397446 | U1283013 | AY750638** | AY750663** | AF1421 | |||
| Zalophus californianus | California sea lion | AF0397456 | Y0852512 | AY750639** | X82310 2 | AY750612** | AY750664** | LSUMZ-044 | |
| Phocidae | Erignathus barbatus | Bearded seal | AF0397426 | AY17004721 | AY17010421 | AY17007721 | AY750665** | AF-1417 | |
| Halichoerus grypus | Grey seal | NC_0016021 | NC_0016021 | NC_0016021 | |||||
| Phoca vitulina | Harbor seal | AY750599** | AY750640** | X823062 | AY750666** | SLZ-920124 | |||
| Procyonidae | Bassaricyon gabbii | Olingo | Y0851012 | X9493111 | |||||
| Potos flavus | Kinkajou | AF0397376 | U783445 | AY750641** | L2187622 | AY750611** | AY750667** | ALG-14904 | |
| Procyon lotor | Common raccoon | AF0397366 | Y0851012 | AY17004621 | X9493011 | AY17007621 | AY750668** | PL-2093 | |
| Ursidae | Ailuropoda melanoleuca | Giant panda | AF0397386 | Y0852112 | AY750642** | X9491811 | AY750669** | NZP-92765 | |
| Helarctos malayanus | Malayan sun bear | U1889920 | |||||||
| Tremartos ornatus | Spectacled bear | AF0397406 | L2188322 | AY17004521 | U2355420 | AY17007521 | AY750670** | PL-959 | |
| Ursus arctos | Grizzly bear (brown bear) | AF0397416 | Y0851912 | AY750643** | X823082 | AY750671** | BZ “Doo” | ||
| Felidae | Acinonyx jubatus | Cheetah | AY750600** | AUD-688 | |||||
| Felis catus | Domestic cat | Y0850312 | NC_00170014 | AB004238 | Z1181119 | ||||
| Felis concolor | Puma | U3349518 | |||||||
| Felis pardalis | Ocelot | AY750601** | U783315 | AY750672** | JMC-298 | ||||
| Felis silvestris | Wild cat | AF0397246 | AY17004221 | AY17010221 | AY17007221 | AY750673** | SDZ-32188 | ||
| Lynx rufus | Bobcat | AY750602** | AY750644** | AY750674** | PL-2025 | ||||
| Panthera leo | Lion | AF0397256 | Y0850512 | AF0530523 | AY750675** | SLZ-076005 | |||
| Panthera tigris | Tiger | AY750603** | Y0850412 | AY750645** | X823012 | AY750676** | PL-942 | ||
| Panthera uncia | Snow leopard | AY750646** | PL-2087 | ||||||
| Herpestidae | Atilax paludinosus | Marsh mongoose | AY750604** | AY750647** | ROM-C1A | ||||
| Crossarchus obscurus | Dark mongoose (kusimanse) | AF0397266 | AY17004121 | AY17010121 | AY17007121 | ||||
| Cynictis pencillata | Yellow mongoose | AY17002421 | AY17004921 | AY17010621 | AY17007921 | ||||
| Galidia elegans (central) | Ring-tailed mongoose | AF0397276 | AY750648** | SMG-6499 | |||||
| Galidia elegans (northern) | Ring-tailed mongoose | AY17002121 | AY17003921 | AY17009921 | AY17006921 | ||||
| Galidia elegans (southern) | Ring-tailed mongoose | AY17002021 | AY17003821 | AY17009821 | AY17006821 | ||||
| Galidictis fasciata | Broad-striped mongoose | AY17002221 | AY17004021 | AY17010021 | AY17007021 | AY750677** | SMG-7554 | ||
| Helogale parvula | Dwarf mongoose | AY750605** | AY750649** | SDZ 30590 | |||||
| Herpestes edwardsii | Indian gray mongoose | AY17002521 | AY17005021 | AY17010721 | AY17008021 | ||||
| Herpestes javanicus | Small Indian mongoose | AY17002621 | Y0850612 | AY17005121 | AY17010821 | AY17008121 | |||
| Liberiictis kuhni | Liberian mongoose | AY750650** | MTZ | ||||||
| Mungos mungo | Banded mongoose | AY17001721 | AY17003521 | AY17009521 | AY17006521 | ||||
| Mungotictis decemlineata | Narrow-striped mongoose | AY17001621 | AY17003421 | AY17009421 | AY17006421 | ||||
| Paracynictis selousi | Gray meerkat (selous' mongoose) | AY750606** | AY750651** | MC-95935 | |||||
| Salanoia concolor | Brown mongoose | AY750607** | AY18700721 | FMNH-33946 | |||||
| Suricata suricatta | Meerkat (suricate) | AY17002821 | D2889916 | AY17005421 | AY17011121 | AY17008421 | |||
| Hyaenidae | Crocuta crocuta | Spotted hyena | AF0397286 | AY17005721 | AY17011421 | AY17008721 | AY750678** | LF “Sargent” | |
| Viverridae | Civettictis civetta | African civet | AY17002321 | AY17004821 | AY17010521 | AY17007821 | |||
| Cryptoprocta ferox | Fossa | AY17001821 | AY17003621 | AY17009621 | AY17006621 | ||||
| Fossa fossa | Fanaloka (Malagasy civet) | AY17001921 | AY17003721 | AY17009721 | AY17006721 | AY750679** | SMG-7539 | ||
| Genetta servalina | Servaline genet | AY17002921 | AY17005821 | AY17011521 | AY17008821 | ||||
| Hemigalus derbyanus | Banded palm civet | AY17002721 | AY17005221 | AY17010921 | AY17008221 | ||||
| Nandinia binotata | African palm civet | AF0397296 | AY17005321 | AY17011021 | AY17008321 | AY750680** | JCK-2623 | ||
| Paradoxurus hermaphroditus | Common palm civet | AF0397306 | AY17005621 | AY17011321 | AY17008621 | ||||
| Viverra tangalunga | Malayan civet | AF0397316 | AY17005521 | AY17011221 | AY17008521 | AY750681** | LRH-4121 | ||
| Family . | Taxon/species . | Common name . | TR . | 12S . | ND2 . | CYTB . | IRBP . | TBG . | Voucher . |
|---|---|---|---|---|---|---|---|---|---|
| Ailuridae | Ailurus fulgens | Red panda | AF0397396 | Y0851112 | AY750613** | X9491911 | AY750608** | AY750652** | NZP 83-671 |
| Canidae | Canis familiaris | Domestic dog | AY750579** | Y0850712 | NC_0020088 | X9492011 | AY750653** | PL2267 | |
| Canis lupus | Gray wolf | AF0397326 | AY17004421 | AY17010321 | AY17007421 | ||||
| Canis rufus | Red wolf | AY750580** | AY750654** | GAG132 | |||||
| Vulpes vulpes | Red fox | AF0397336 | Y0850712 | AY750614** | X9492911 | AY750655** | PL2023 | ||
| Mustelidae | Aonyx capensis | Short clawed or clawless otter | AY750615** | AF0571182 | PL4663 | ||||
| Conepatus mesoleucus | Hog-nosed skunk | AY750581** | U783265 | AY750616** | AY750656** | NK 43633 | |||
| Eira barbara | Tayra | AY750582** | AY750617** | SDZ 588414 | |||||
| Enhydra lutris | Sea otter | AY750583** | Y0851212 | AY750618** | AF0571209 | AB082978‡17 | AY750657** | SA “Nuka” | |
| Gulo gulo | Wolverine | AY750584** | U783335 | AY750619** | X9492111 | AB082962‡17 | BZ-47-F4 | ||
| Ictonyx striatus | Zorilla (African polecat) | AY750585** | U783345 | BZ-880085 | |||||
| Lontra canadensis | North American river otter | AY750586** | U783355 | AY750620** | AF0571219 | AY750620** | PL 2068 | ||
| Lontra longicaudis | Neotropical river otter | AF0397346 | AY750621** | AF0571239 | ALG-14988 | ||||
| Martes americana | American marten | AY750587** | U783365 | AY750622** | AF0571309 | AB082963‡17 | BZ-47-C8 | ||
| Martes flavigula | Yellow-throated marten | AY750588** | AY750623** | AB01236310 | AB082964‡17 | SDZ-32799 | |||
| Martes pennanti | Fisher | AY750589** | AY750624** | AF0571319 | PL-4273 | ||||
| Mephitis mephitis | Striped skunk | AF3069487 | Y0851712 | AY750625** | X9492711 | AY750609** | AY750659** | NK 43631 | |
| Mustela erminea | Ermine (stoat) | AY750590** | AY750626** | AB02610115 | AB082969‡17 | LJB-2435 | |||
| Mustela frenata | Long-tailed weasel | AF0397356 | U783395 | AY750627** | AF0685474 | AY750660** | PL-4656 | ||
| Mustela lutreola | European mink | AY750591** | AY750628** | AB02610515 | AB082972‡17 | PL-3441 | |||
| Mustela nivalis | European common weasel | AY750592** | Y0851512 | AY750629** | AB02610615 | AB082973‡17 | SDZ5001 | ||
| Mustela putorius | European polecat | AY750593** | Y0851612 | AY750630** | AB02610715 | AB082975‡17 | SDZ-33221 | ||
| Mustela sibirica | Siberian weasel (kolinsky) | AY750594** | AY750631** | AB02610815 | SDZ-32108 | ||||
| Mustela vision | American mink | AY750595** | Y0851412 | AY750632** | AB02610915 | AB082977‡17 | LJB-2124 | ||
| Mydaus spp. | Stink badger | AY750596** | U783425 | AY750633** | JDW | ||||
| Poecilogale albinucha | African striped weasel | AY750597** | AY750634** | JCK2891 | |||||
| Spilogale putorius | Spotted skunk | AF3069497 | U783465 | AY750635** | X9492811 | AY750610** | AY750661** | NK 43632 | |
| Taxidea taxus | American badger | AY750598** | U783475 | AY750636** | AF0571329 | PL2542 | |||
| Odobenidae | Odobenus rosmarus | Walrus | AF0397436 | U783435 | AY750637** | X822992 | AY750662** | BZ910053 | |
| Otariidae | Arctocephalus gazella | Southern fur seal | Y0852612 | X822929 | |||||
| Callorhinus ursinus | Northern fur seal | AF0397446 | U1283013 | AY750638** | AY750663** | AF1421 | |||
| Zalophus californianus | California sea lion | AF0397456 | Y0852512 | AY750639** | X82310 2 | AY750612** | AY750664** | LSUMZ-044 | |
| Phocidae | Erignathus barbatus | Bearded seal | AF0397426 | AY17004721 | AY17010421 | AY17007721 | AY750665** | AF-1417 | |
| Halichoerus grypus | Grey seal | NC_0016021 | NC_0016021 | NC_0016021 | |||||
| Phoca vitulina | Harbor seal | AY750599** | AY750640** | X823062 | AY750666** | SLZ-920124 | |||
| Procyonidae | Bassaricyon gabbii | Olingo | Y0851012 | X9493111 | |||||
| Potos flavus | Kinkajou | AF0397376 | U783445 | AY750641** | L2187622 | AY750611** | AY750667** | ALG-14904 | |
| Procyon lotor | Common raccoon | AF0397366 | Y0851012 | AY17004621 | X9493011 | AY17007621 | AY750668** | PL-2093 | |
| Ursidae | Ailuropoda melanoleuca | Giant panda | AF0397386 | Y0852112 | AY750642** | X9491811 | AY750669** | NZP-92765 | |
| Helarctos malayanus | Malayan sun bear | U1889920 | |||||||
| Tremartos ornatus | Spectacled bear | AF0397406 | L2188322 | AY17004521 | U2355420 | AY17007521 | AY750670** | PL-959 | |
| Ursus arctos | Grizzly bear (brown bear) | AF0397416 | Y0851912 | AY750643** | X823082 | AY750671** | BZ “Doo” | ||
| Felidae | Acinonyx jubatus | Cheetah | AY750600** | AUD-688 | |||||
| Felis catus | Domestic cat | Y0850312 | NC_00170014 | AB004238 | Z1181119 | ||||
| Felis concolor | Puma | U3349518 | |||||||
| Felis pardalis | Ocelot | AY750601** | U783315 | AY750672** | JMC-298 | ||||
| Felis silvestris | Wild cat | AF0397246 | AY17004221 | AY17010221 | AY17007221 | AY750673** | SDZ-32188 | ||
| Lynx rufus | Bobcat | AY750602** | AY750644** | AY750674** | PL-2025 | ||||
| Panthera leo | Lion | AF0397256 | Y0850512 | AF0530523 | AY750675** | SLZ-076005 | |||
| Panthera tigris | Tiger | AY750603** | Y0850412 | AY750645** | X823012 | AY750676** | PL-942 | ||
| Panthera uncia | Snow leopard | AY750646** | PL-2087 | ||||||
| Herpestidae | Atilax paludinosus | Marsh mongoose | AY750604** | AY750647** | ROM-C1A | ||||
| Crossarchus obscurus | Dark mongoose (kusimanse) | AF0397266 | AY17004121 | AY17010121 | AY17007121 | ||||
| Cynictis pencillata | Yellow mongoose | AY17002421 | AY17004921 | AY17010621 | AY17007921 | ||||
| Galidia elegans (central) | Ring-tailed mongoose | AF0397276 | AY750648** | SMG-6499 | |||||
| Galidia elegans (northern) | Ring-tailed mongoose | AY17002121 | AY17003921 | AY17009921 | AY17006921 | ||||
| Galidia elegans (southern) | Ring-tailed mongoose | AY17002021 | AY17003821 | AY17009821 | AY17006821 | ||||
| Galidictis fasciata | Broad-striped mongoose | AY17002221 | AY17004021 | AY17010021 | AY17007021 | AY750677** | SMG-7554 | ||
| Helogale parvula | Dwarf mongoose | AY750605** | AY750649** | SDZ 30590 | |||||
| Herpestes edwardsii | Indian gray mongoose | AY17002521 | AY17005021 | AY17010721 | AY17008021 | ||||
| Herpestes javanicus | Small Indian mongoose | AY17002621 | Y0850612 | AY17005121 | AY17010821 | AY17008121 | |||
| Liberiictis kuhni | Liberian mongoose | AY750650** | MTZ | ||||||
| Mungos mungo | Banded mongoose | AY17001721 | AY17003521 | AY17009521 | AY17006521 | ||||
| Mungotictis decemlineata | Narrow-striped mongoose | AY17001621 | AY17003421 | AY17009421 | AY17006421 | ||||
| Paracynictis selousi | Gray meerkat (selous' mongoose) | AY750606** | AY750651** | MC-95935 | |||||
| Salanoia concolor | Brown mongoose | AY750607** | AY18700721 | FMNH-33946 | |||||
| Suricata suricatta | Meerkat (suricate) | AY17002821 | D2889916 | AY17005421 | AY17011121 | AY17008421 | |||
| Hyaenidae | Crocuta crocuta | Spotted hyena | AF0397286 | AY17005721 | AY17011421 | AY17008721 | AY750678** | LF “Sargent” | |
| Viverridae | Civettictis civetta | African civet | AY17002321 | AY17004821 | AY17010521 | AY17007821 | |||
| Cryptoprocta ferox | Fossa | AY17001821 | AY17003621 | AY17009621 | AY17006621 | ||||
| Fossa fossa | Fanaloka (Malagasy civet) | AY17001921 | AY17003721 | AY17009721 | AY17006721 | AY750679** | SMG-7539 | ||
| Genetta servalina | Servaline genet | AY17002921 | AY17005821 | AY17011521 | AY17008821 | ||||
| Hemigalus derbyanus | Banded palm civet | AY17002721 | AY17005221 | AY17010921 | AY17008221 | ||||
| Nandinia binotata | African palm civet | AF0397296 | AY17005321 | AY17011021 | AY17008321 | AY750680** | JCK-2623 | ||
| Paradoxurus hermaphroditus | Common palm civet | AF0397306 | AY17005621 | AY17011321 | AY17008621 | ||||
| Viverra tangalunga | Malayan civet | AF0397316 | AY17005521 | AY17011221 | AY17008521 | AY750681** | LRH-4121 | ||
**DNA Sequence new to this study.
‡IRBP sequence data from Sato et al. (2003) added for replicate analyses only (see text).
Vouchers for new sequence data are given with the following abbreviations for institutions: (AF) = University of Alaska, Fairbanks Museum; (ALG) = USFWS; (AUD) = Audubon Zoo; (BZ) = Brookfield Zoo; (JWD, NK) = Museum of Southwestern Biology University of New Mexico, Albuquerque; (LF) = University of California, Berkeley; (JCK LRH SMG PL MC GAG) = Field Museum of Natural History; (LSUMZ, JCS, JMC, LJB) = Louisiana State University, Museum of Zoology; (MTZ) = Metro Toronto Zoo; (NZP) = National Zoological Park, Washington D.C.; (ROM) = University of Toronto, Royal Ontario Museum; (SA) = Shedd Aquarium; (SDZ) = San Diego Zoo; (SLZ) = St. Louis Zoo.
REFERENCES
Author notes
Current Address: Division of Paleontology, American Museum of Natural History, Central Park West at 79th Street, New York, NY, 10024, USA
Current Address: P.O. Box 637, Maple Valley, Washington, 98038, USA